Part II: Pragmatic Solutions and Best Practices
Save time, reduce errors, and work more efficiently in teams
September 8, 2025
Welcome to Part II

Pragmatic Solutions and Best Practices
Motivation
- Write clear, reliable R scripts for real projects (not package dev)
- Save time on maintenance
- Reduce errors
- Make teamwork smoother and simplify handovers
- Leave with online rescources that help to deepen the topic
Who is this for?
- Statisticians writing R scripts, e.g., for clinical trial planning & analysis
- People who collaborate in teams and hand over code
- You don’t need to be a software engineer to benefit
The reality we all know
- Tight timelines, changing specs, handovers
- Old scripts reused under pressure
- “I’ll clean this later” — later never comes
- Result: time loss, bugs, stress

GitHub From the Beginning

GitHub tells the whole story of your project
GitHub as your business card
- GitHub as your business card and career showcase
- Who would you hire?
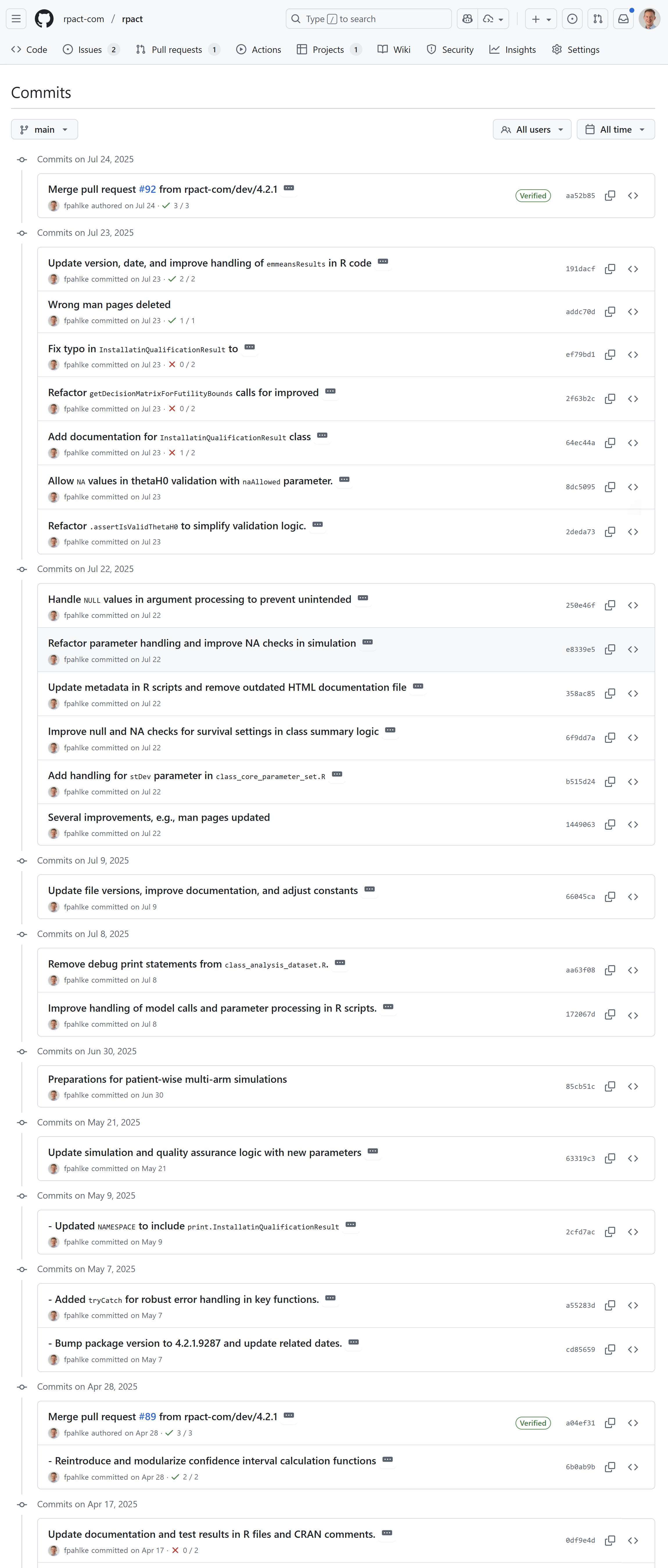
Fictive example: Clinical trial analysis

Let’s use GitHub
Create a new repository on GitHub, clone it to your local machine, and add a README.md file with a brief description of your project.
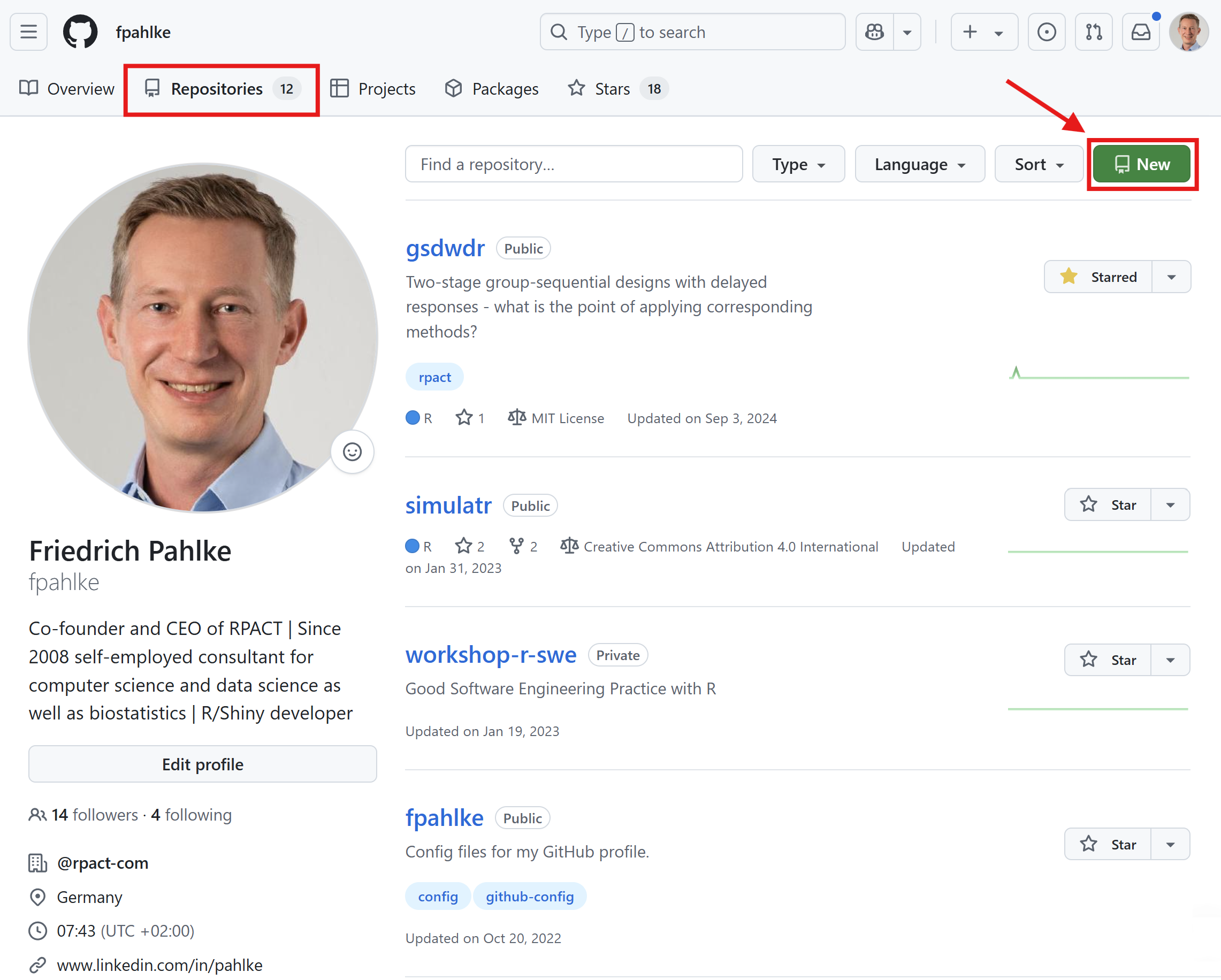

TortoiseGit
Windows context menu for GitHub: TortoiseGit

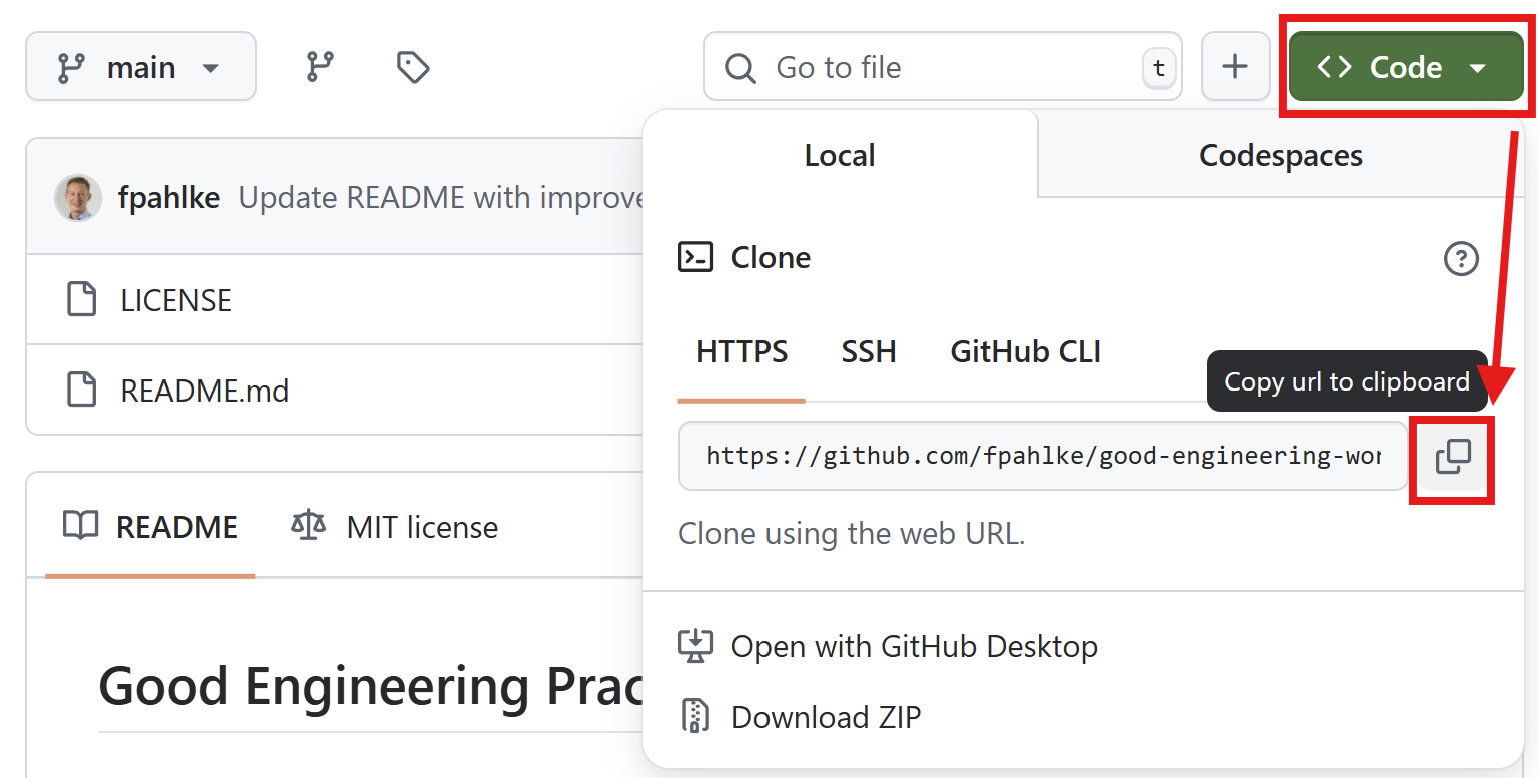
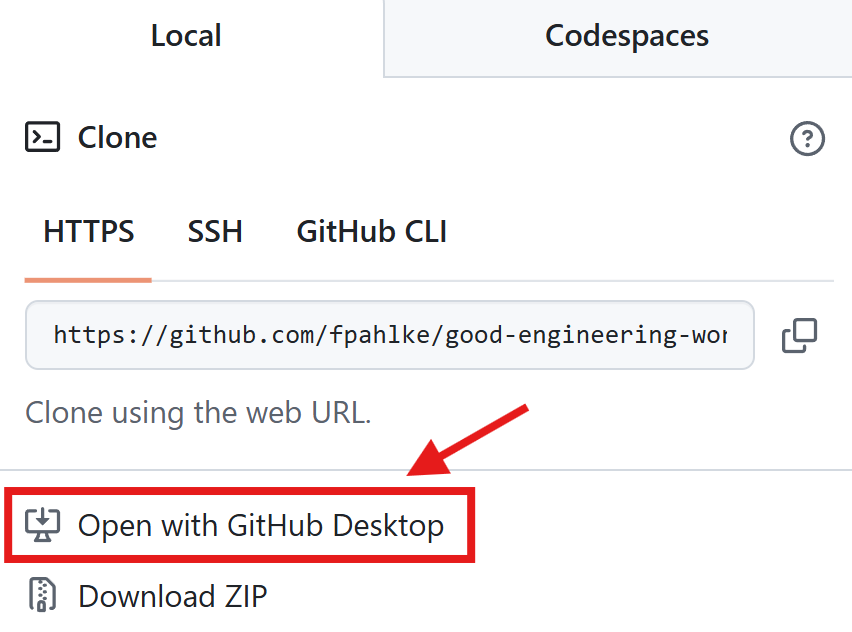
GitHub Desktop
GitHub Desktop App
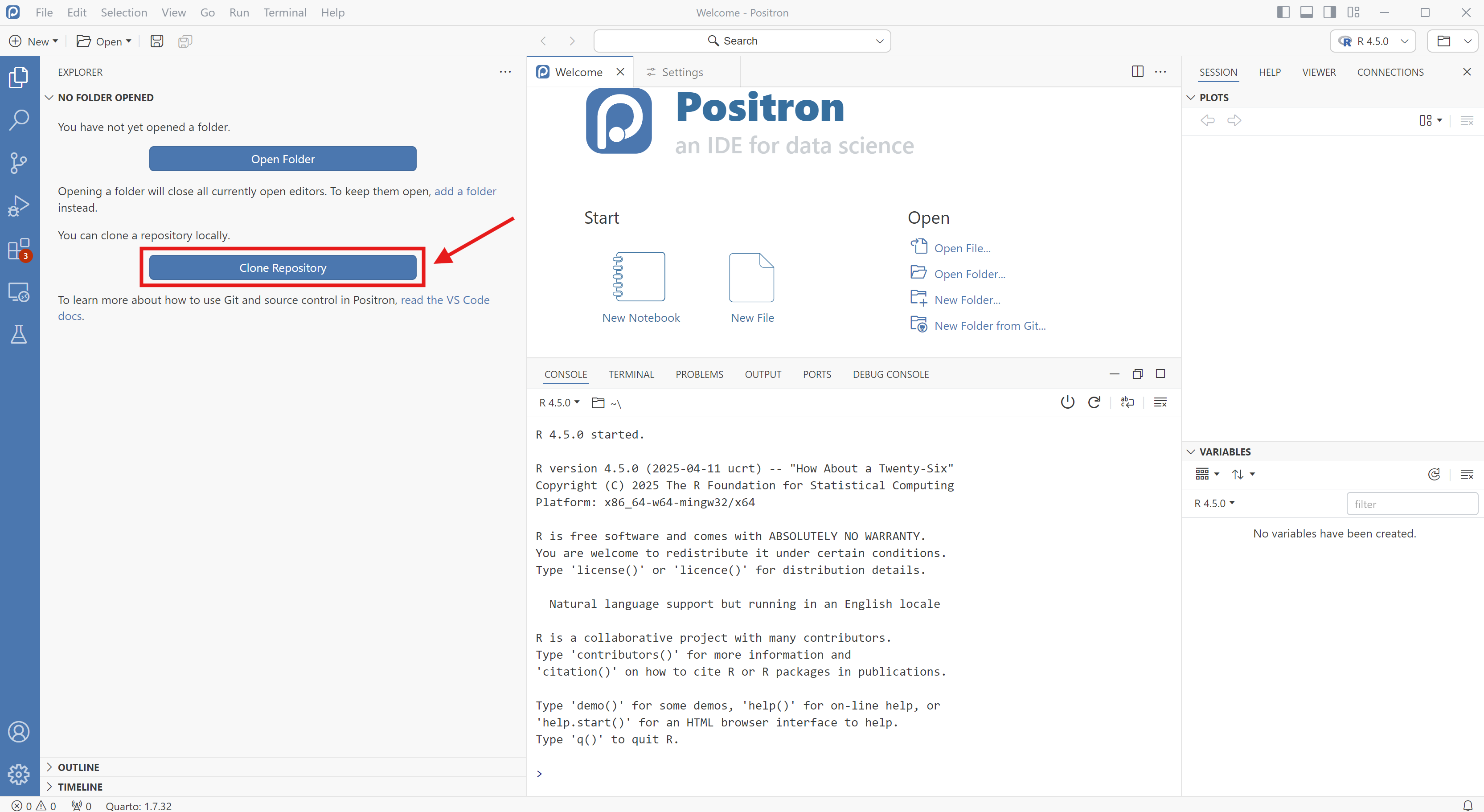
Other GitHub Apps
Many other ways to use GitHub: Eclipse, Positron, VS Code, RStudio, …
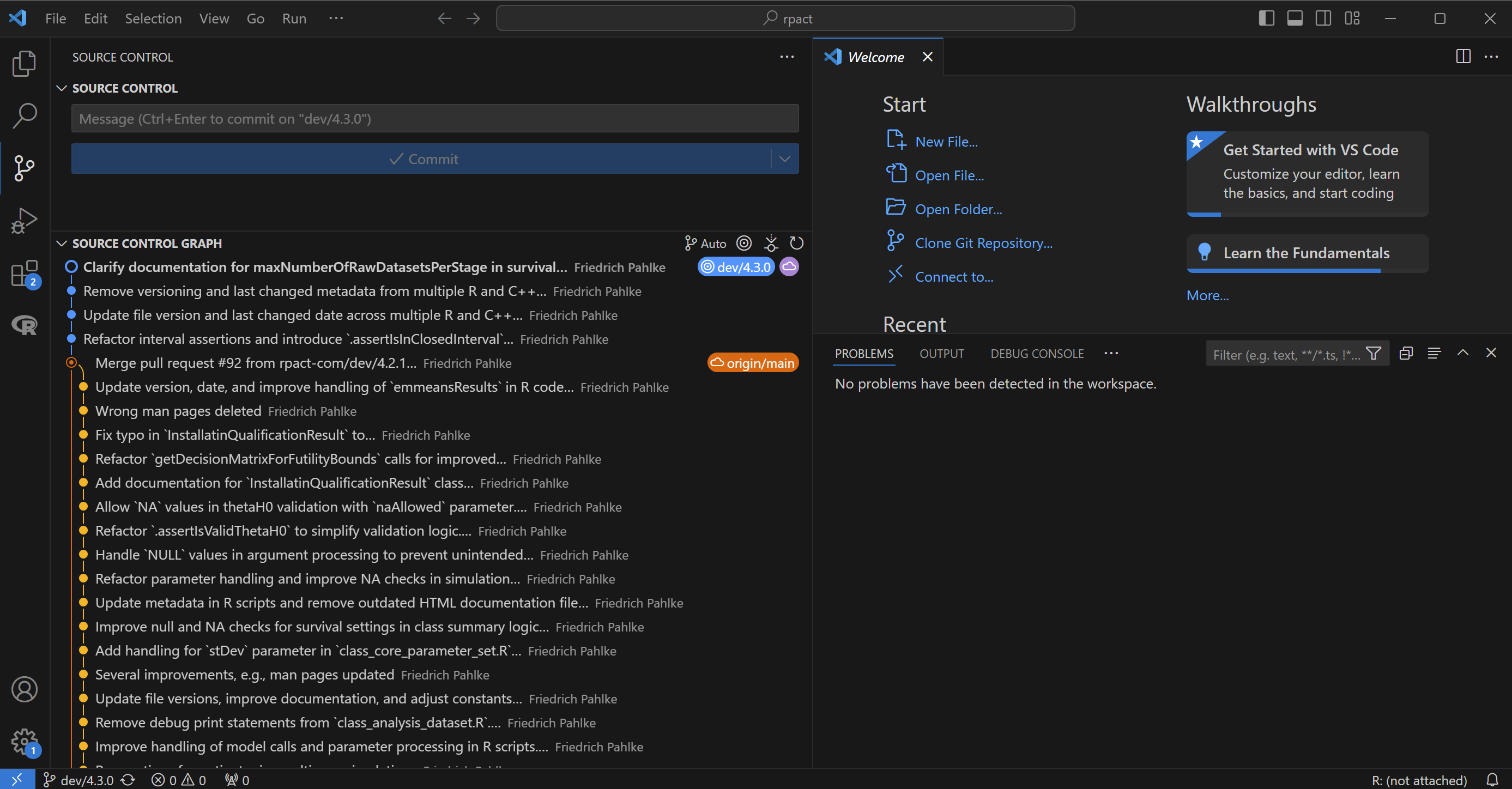
One branch per developer
- Each developer works on his own branch(es)
- Each branch is merged into
mainonly after review mainis always stable, ready for production- Let’s say we have 2 developers:
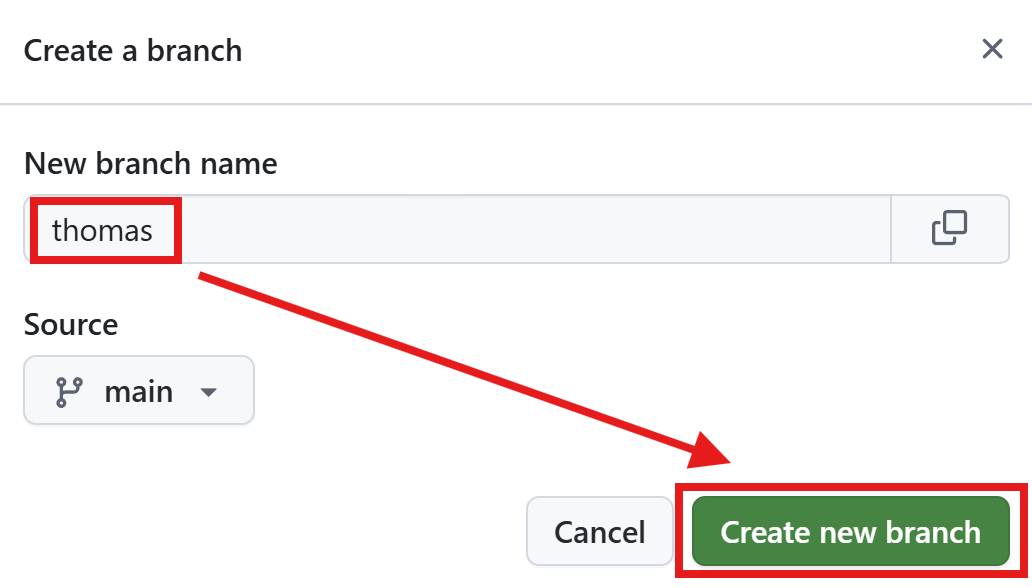
thomas,friedrich - Friedrich’s job is it to program a new R script and learn how to apply the principles and patterns of this workshop
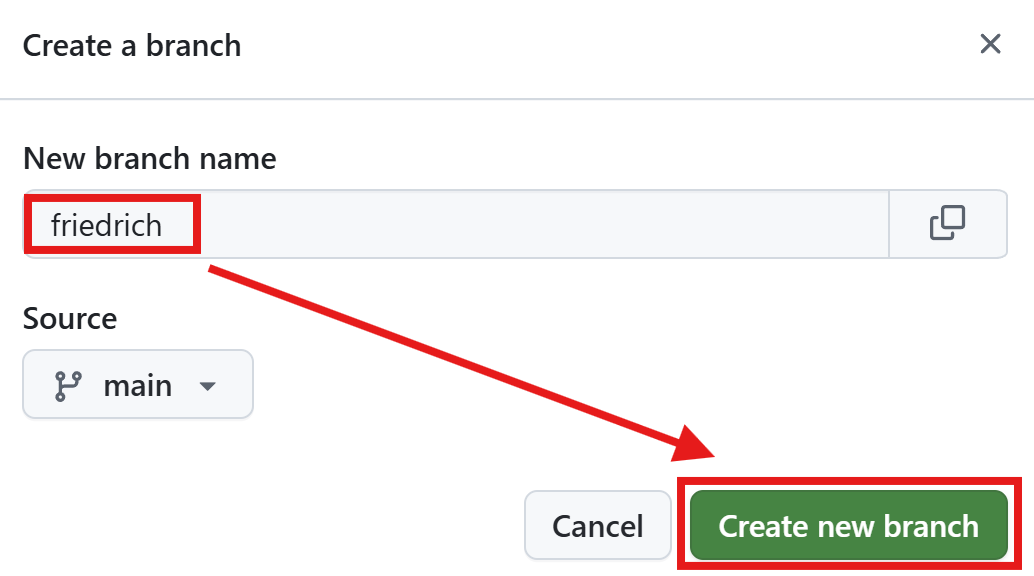
- Thomas is responsible for the code review and instruction of Friedrich
Let’s create the two branches

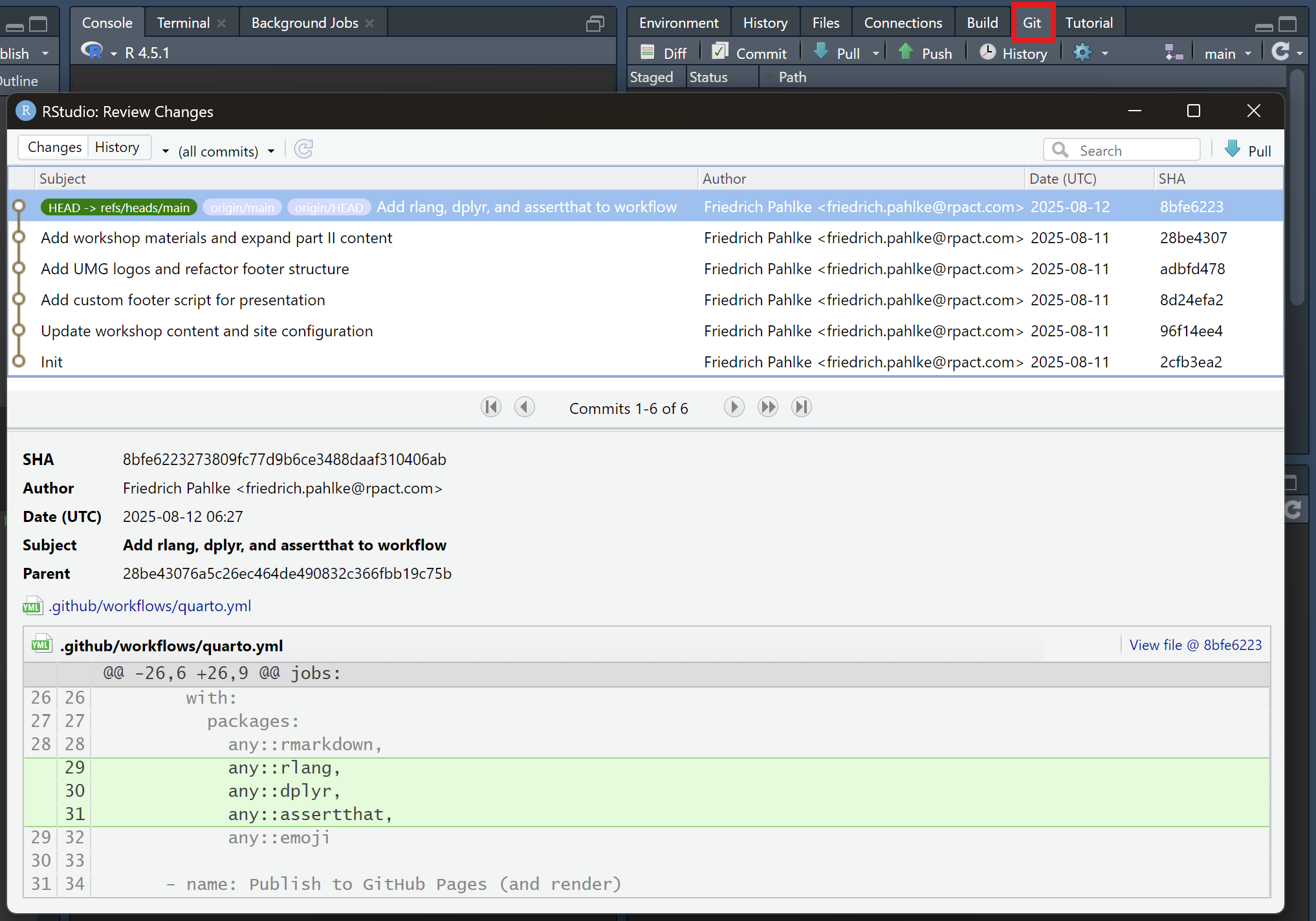
Branches
github.com/fpahlke/good-engineering-workshop-demo/branche
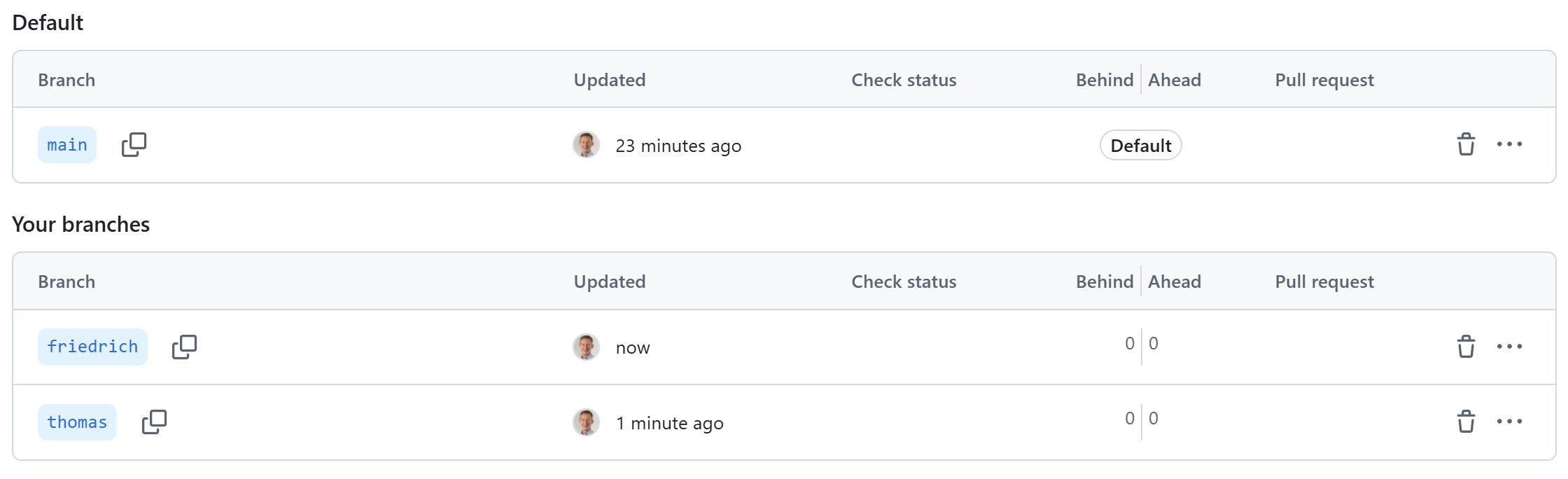
GitHub branches overview page
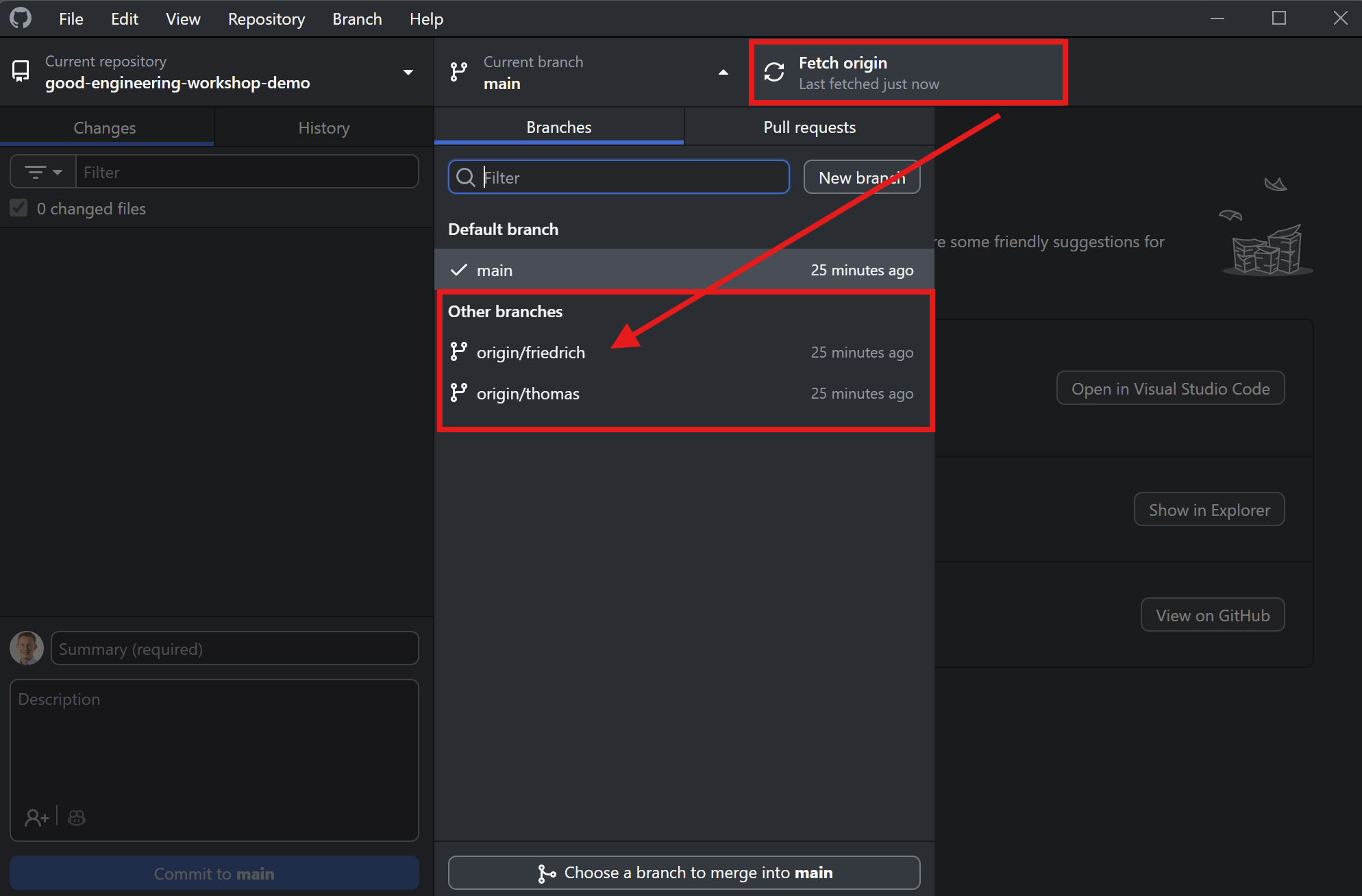
Let’s clone the repository with GitHub Desktop
Fetch origin (update local information) and then select branch friedrich.
Add a new R script and data file
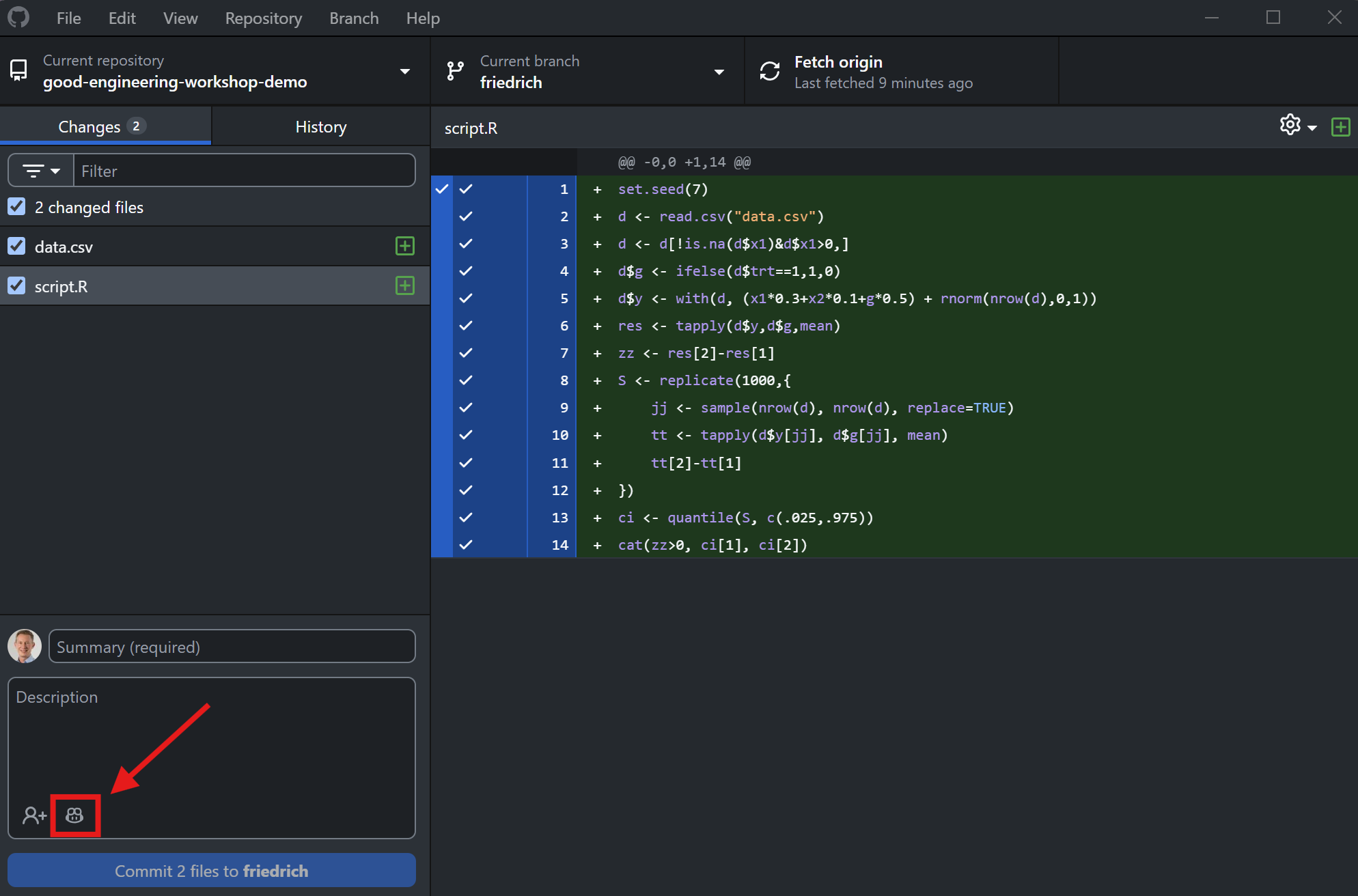
Add a new R script and data file
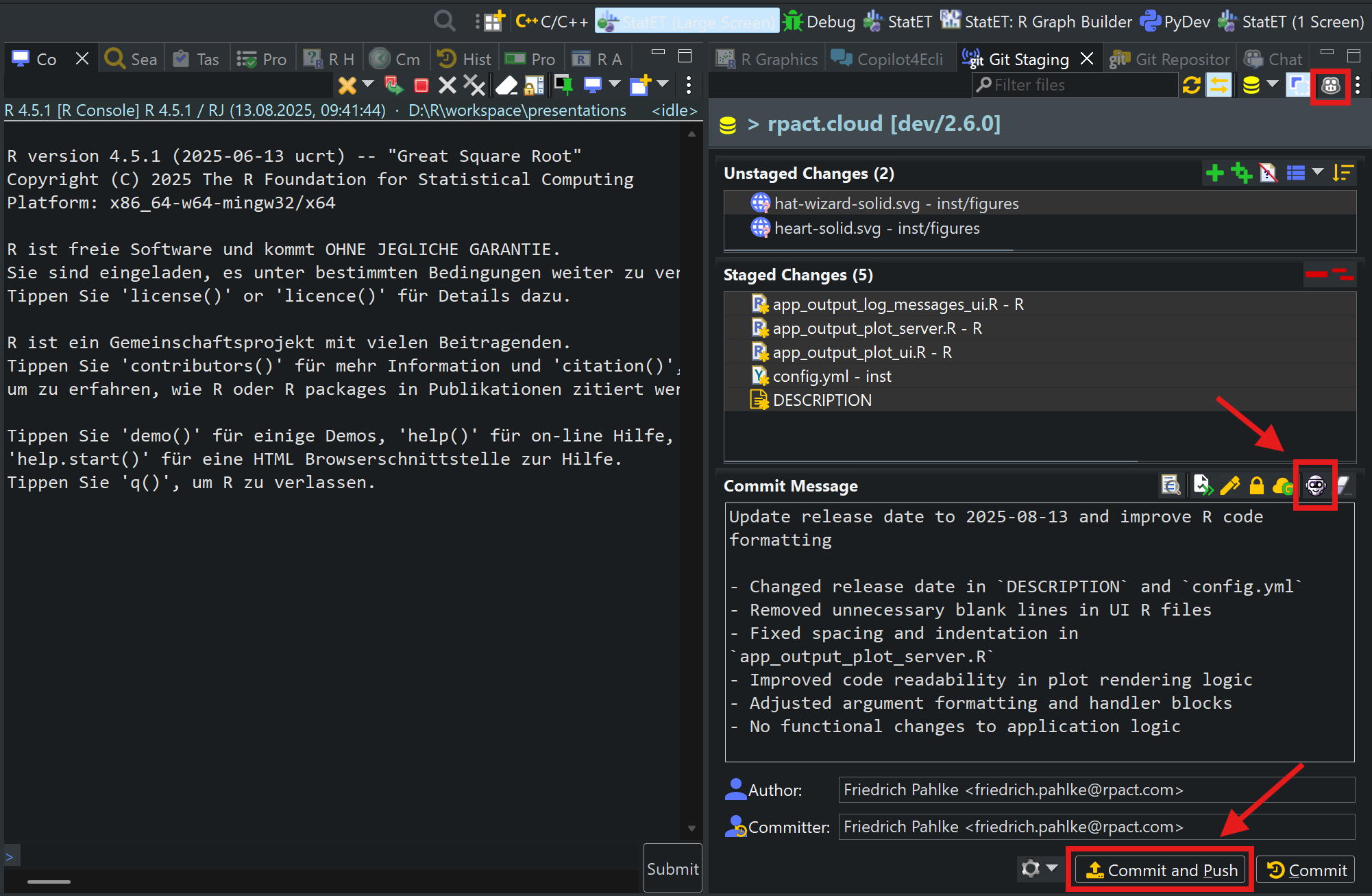
Use Copilot to write the commit message
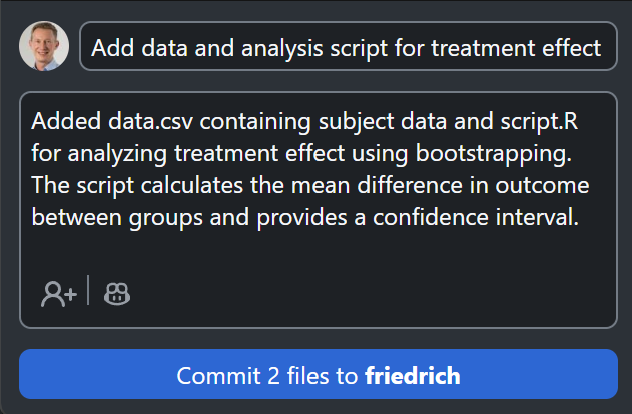
Push the changes to GitHub
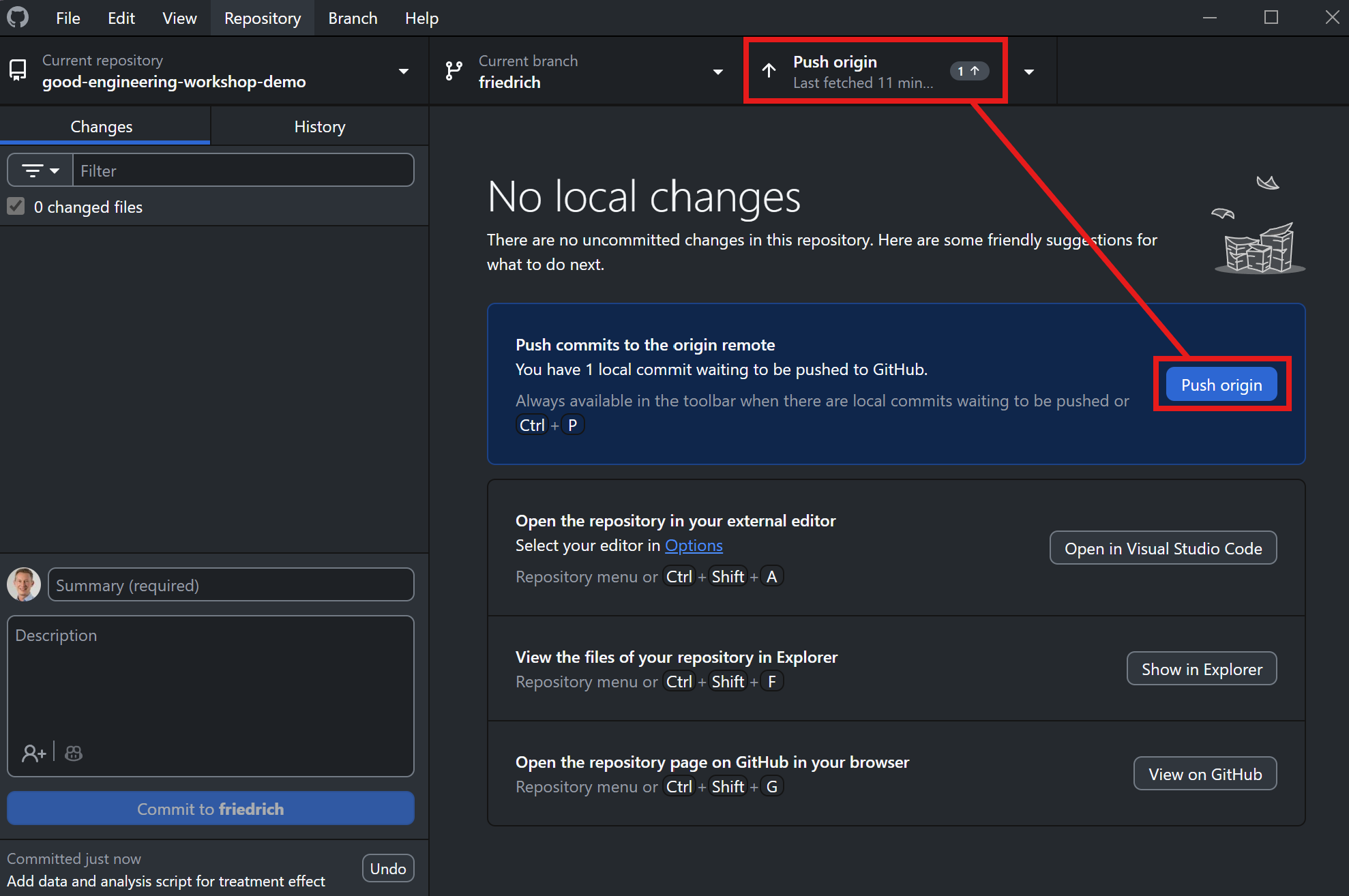
Push the changes to GitHub
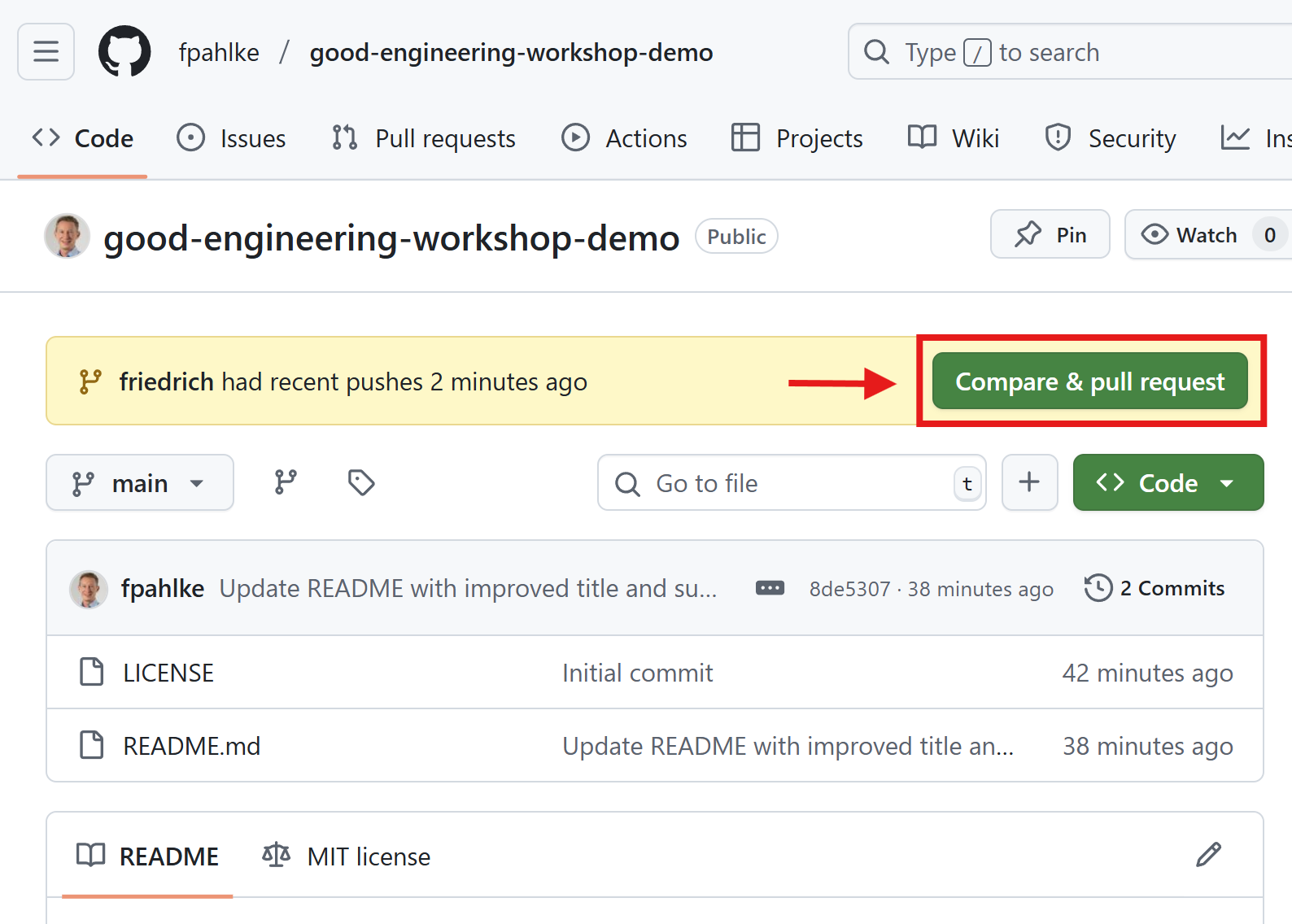
Create a pull request
Use Copilot for the pull request description and review of the changes.
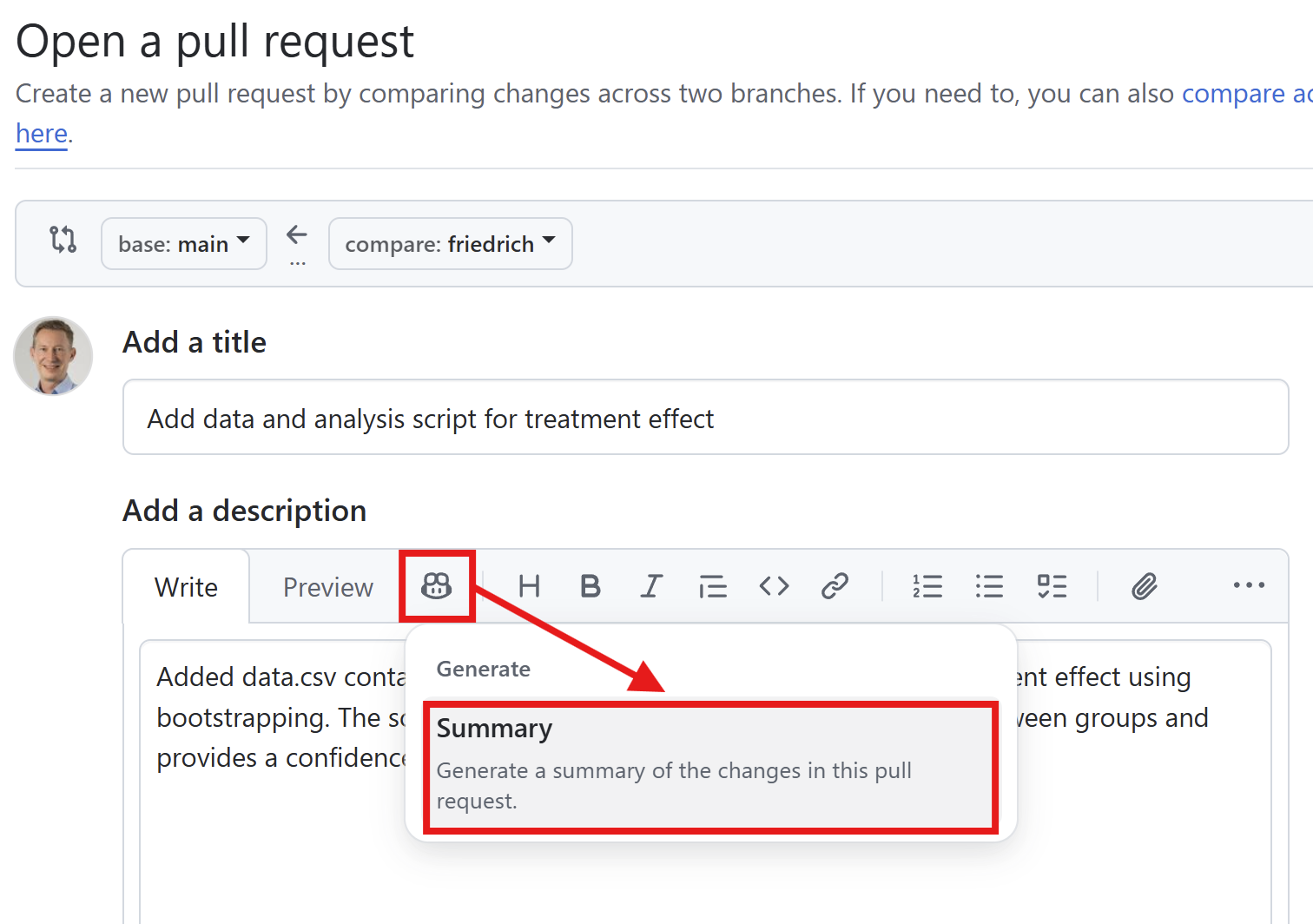
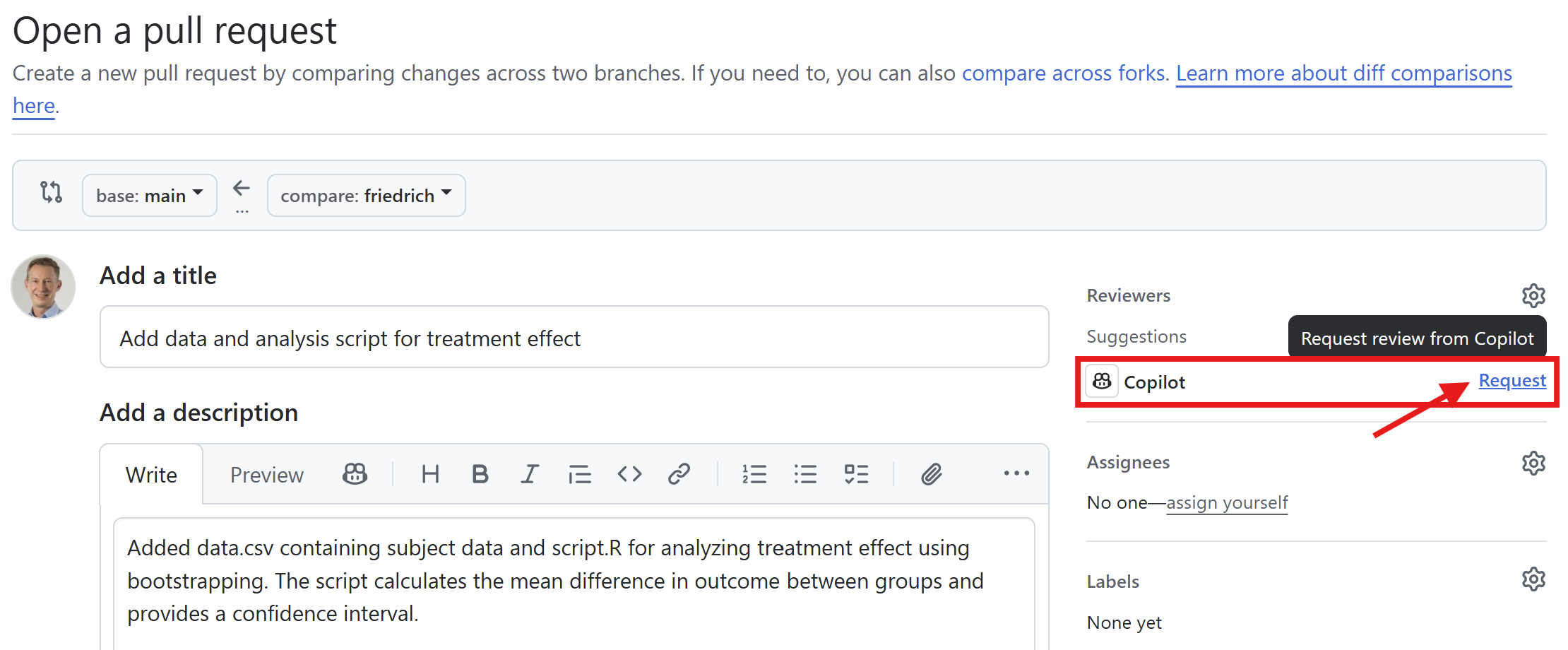
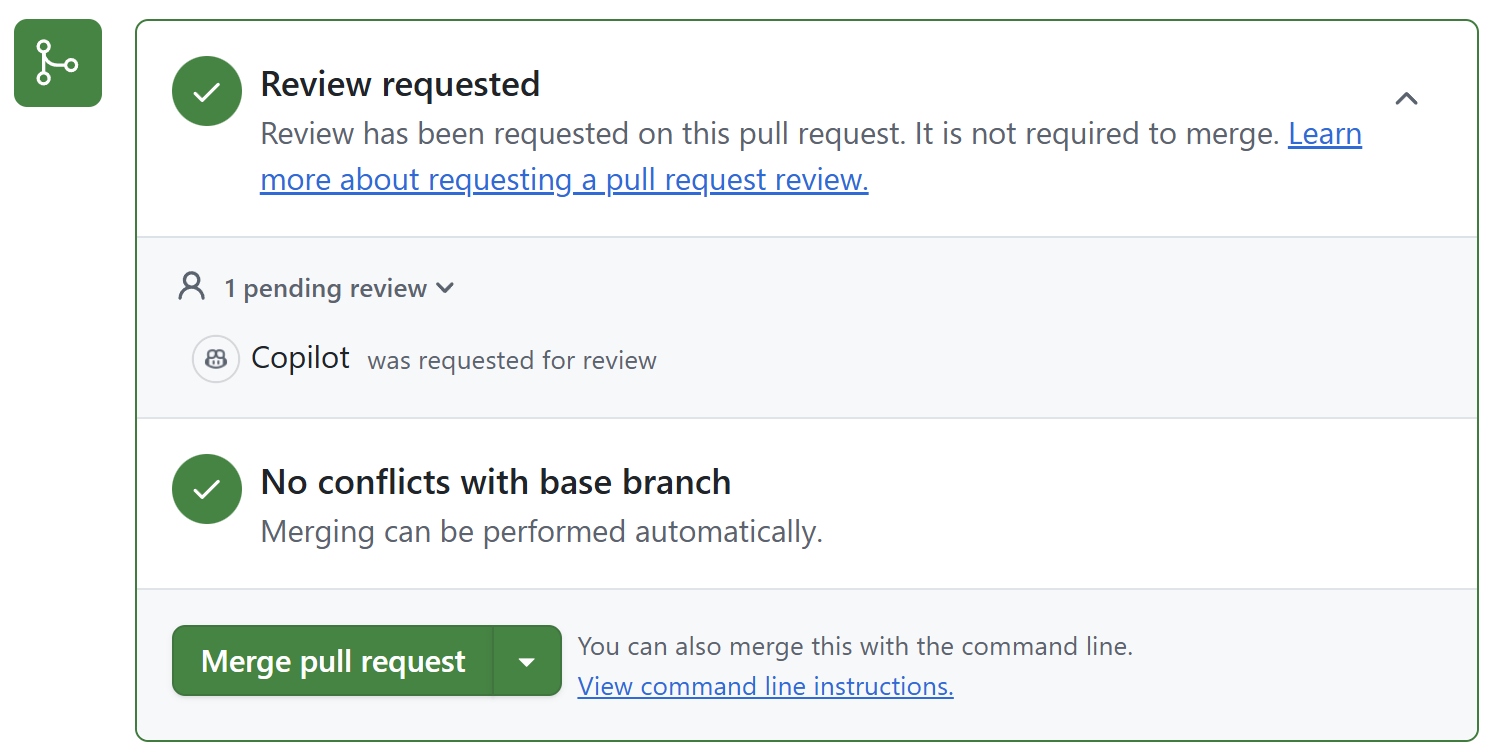
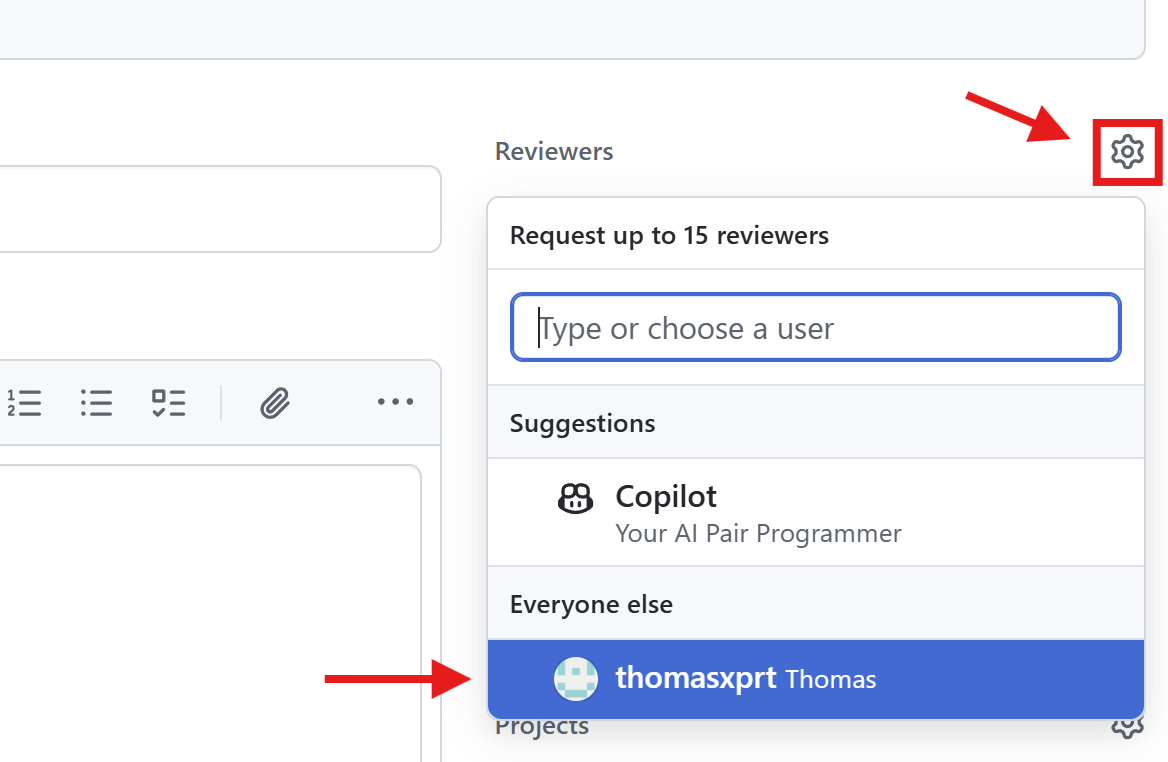
Add additional reviewers
First invite Thomas as collaborator.
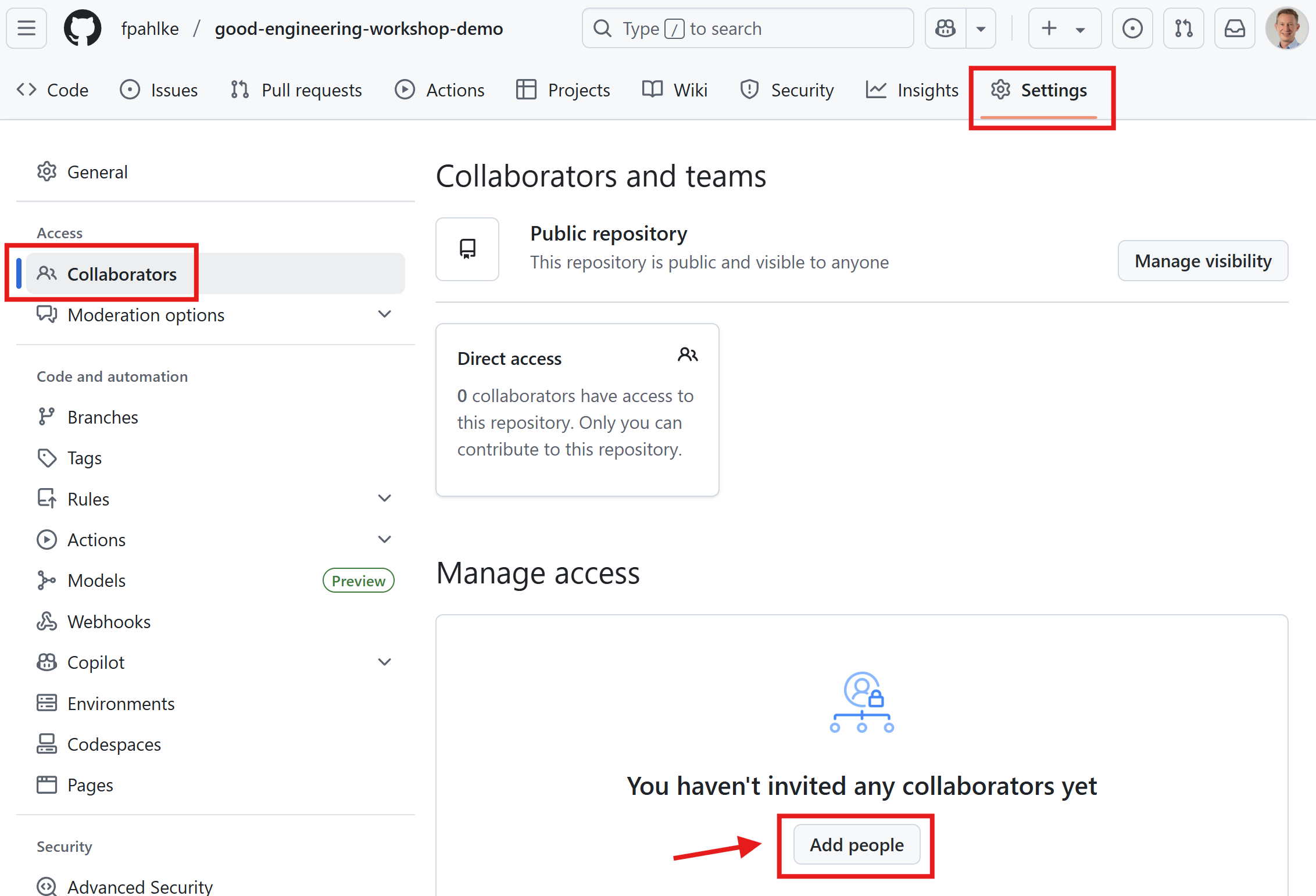
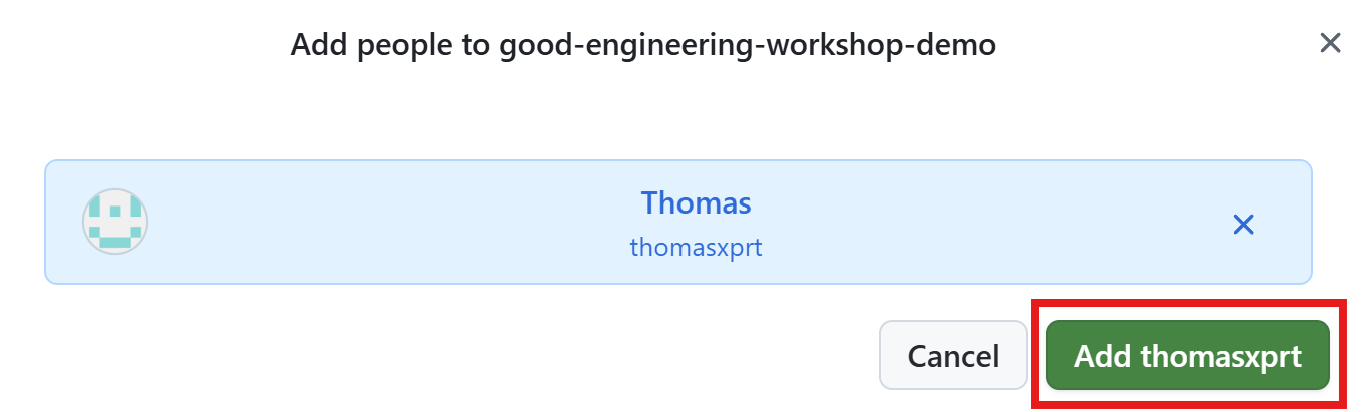
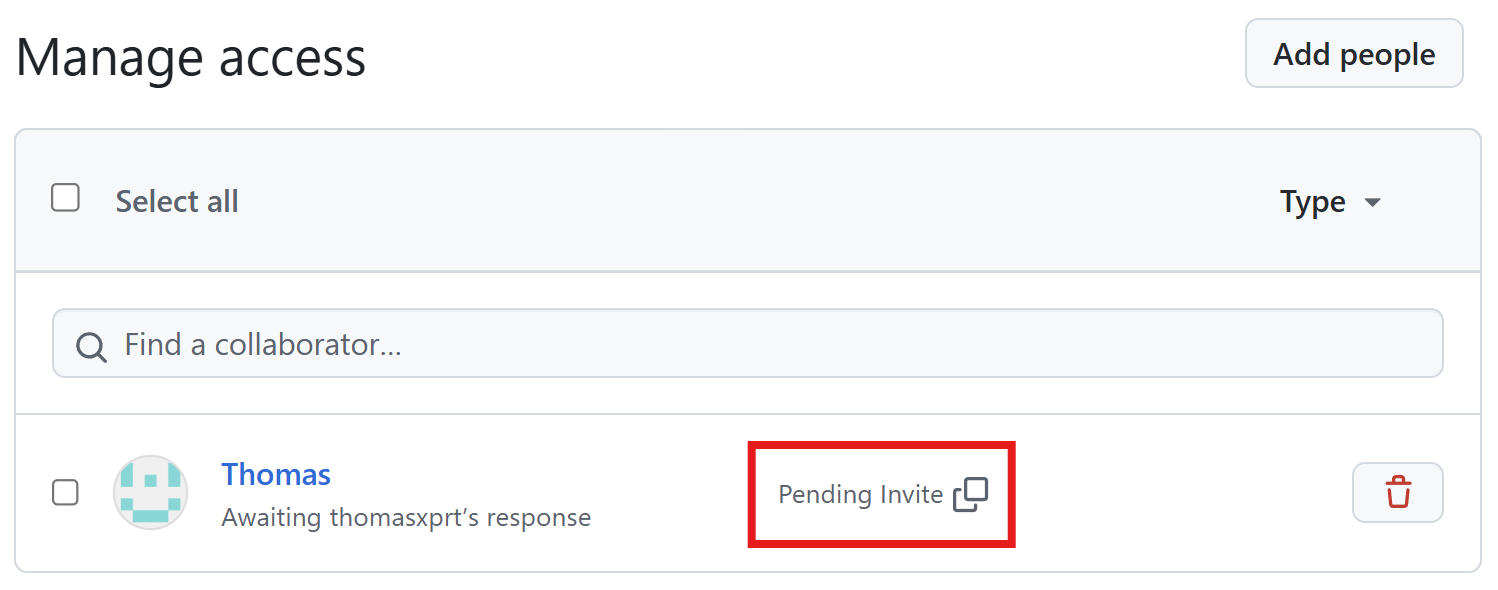
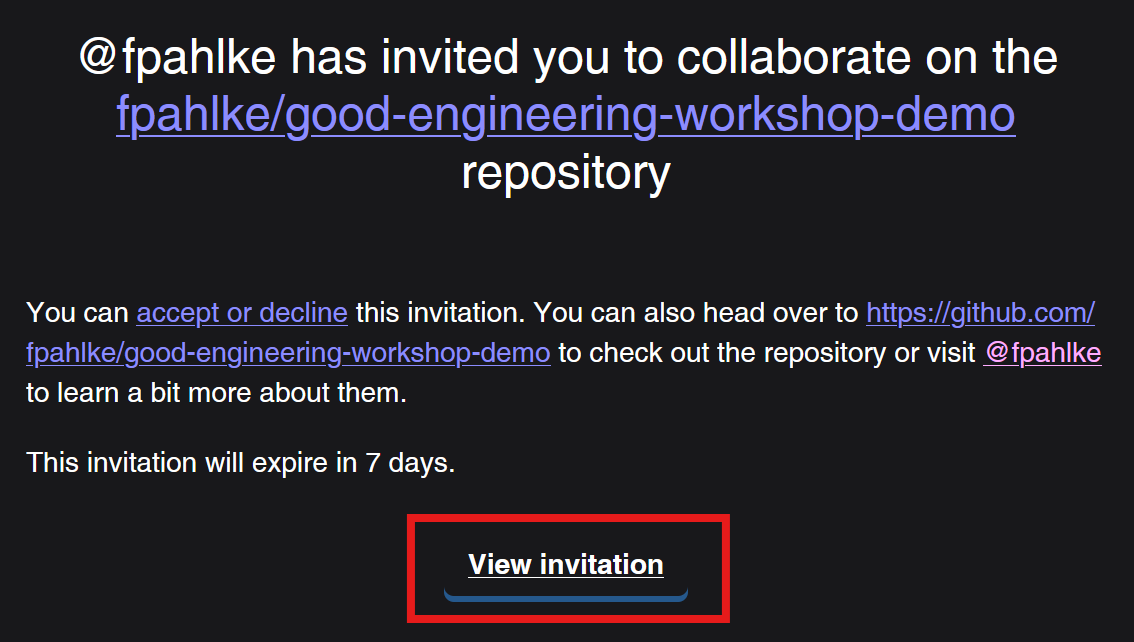
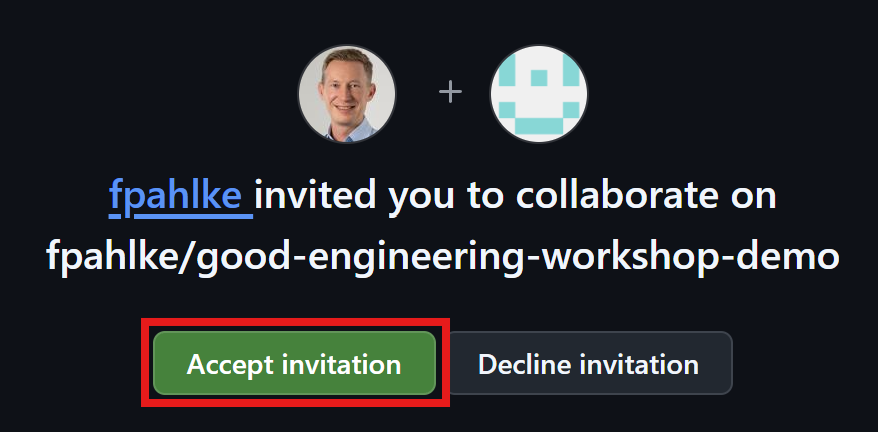
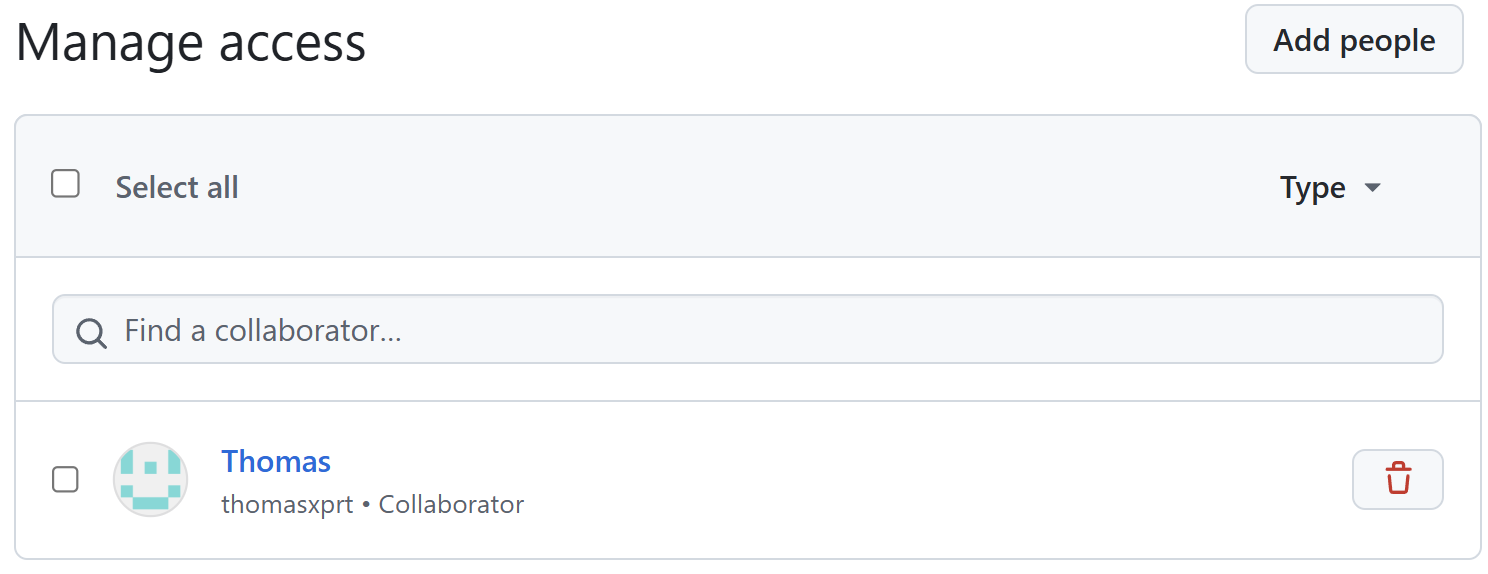
Add additional reviewers (cont’d)
Then add Thomas as reviewer.


Thomas reviews the pull request
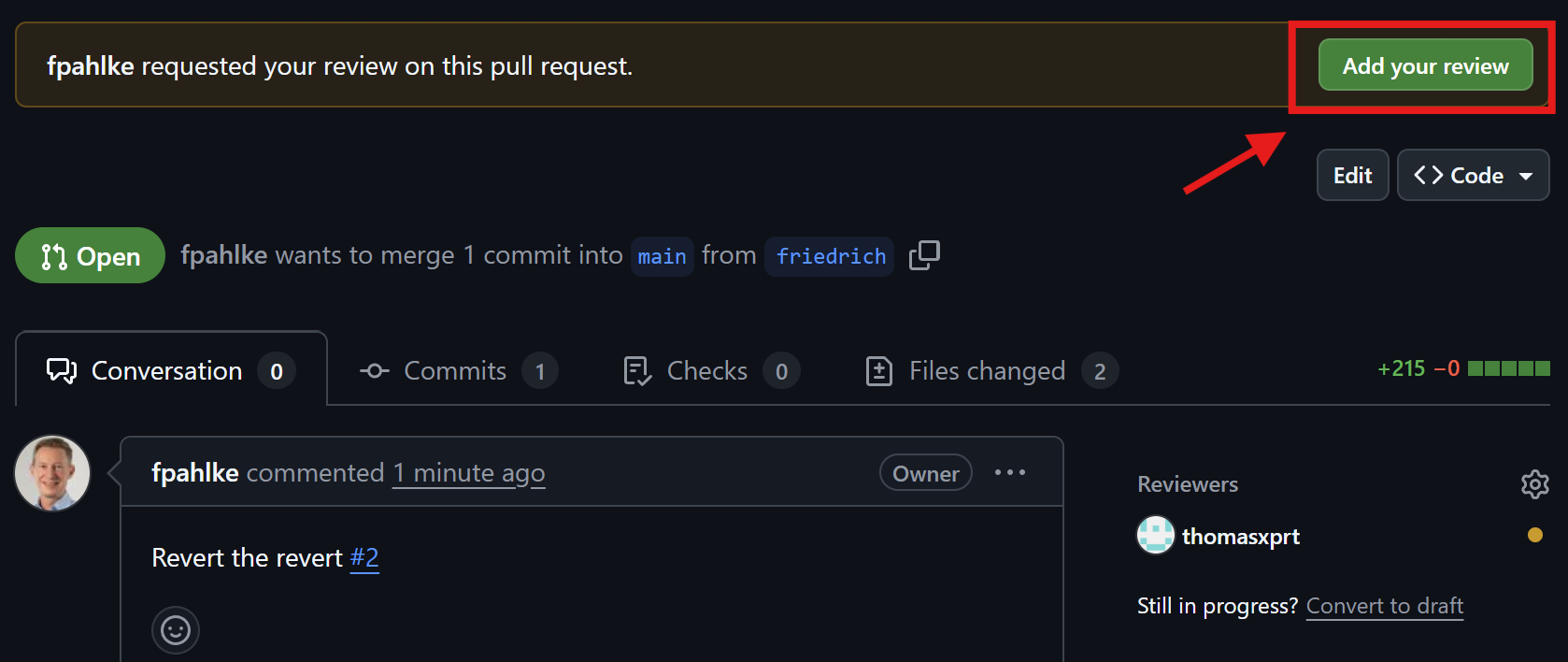
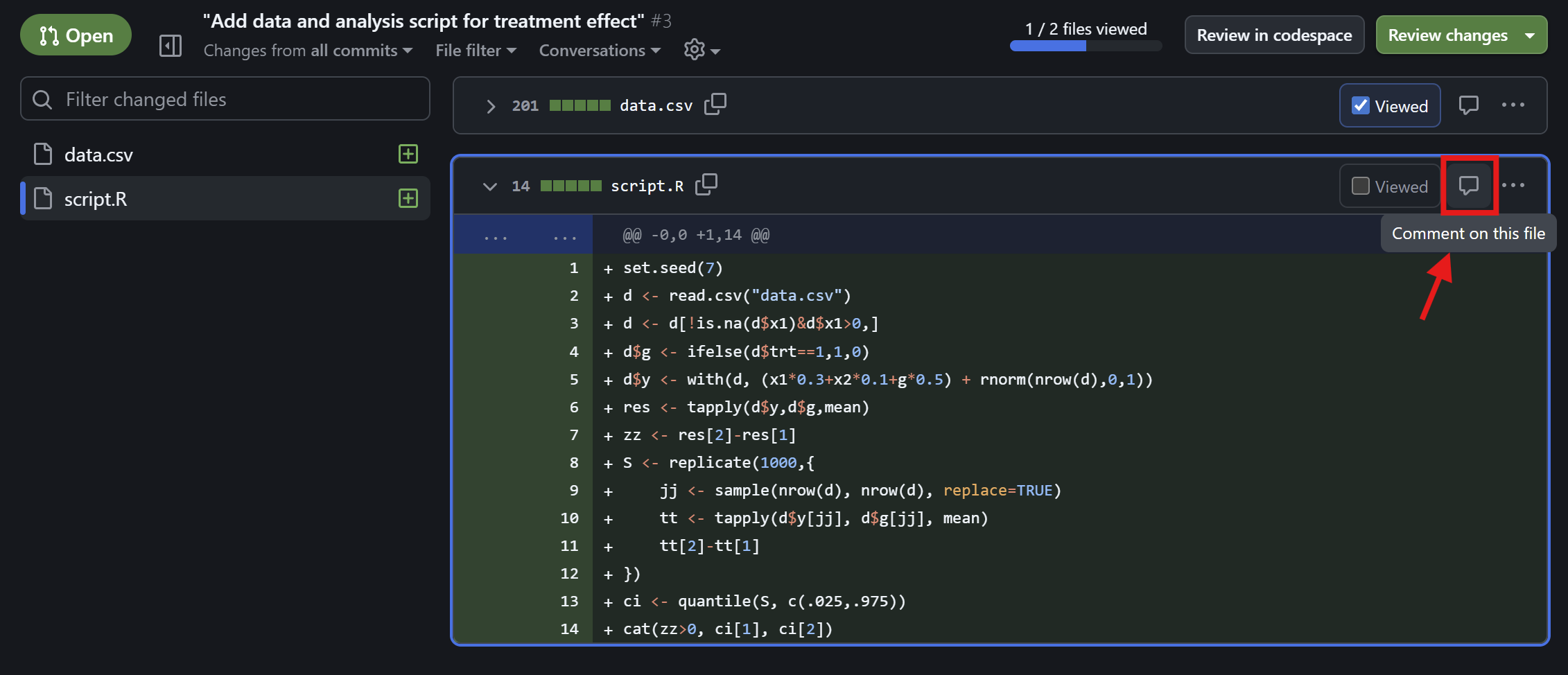

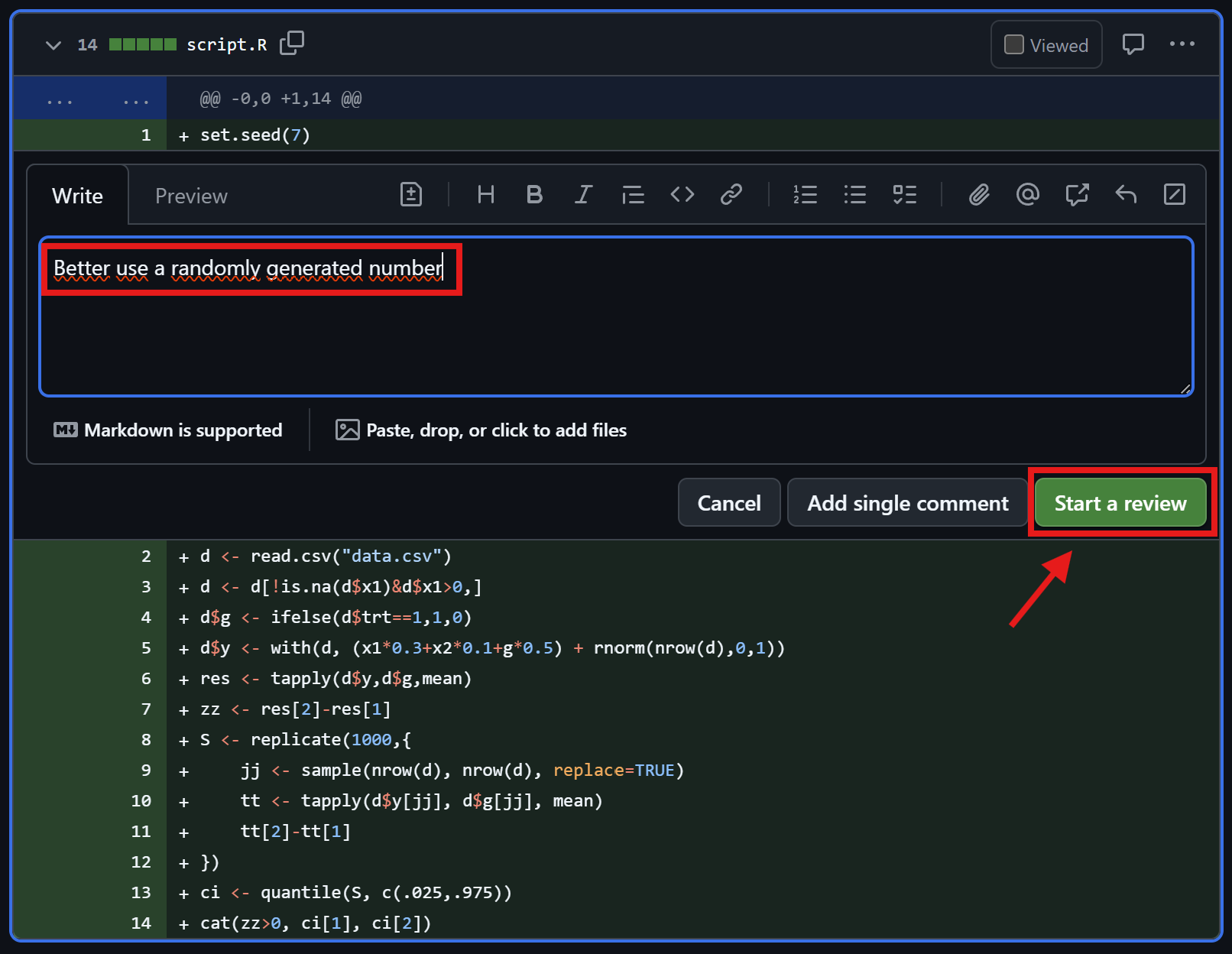
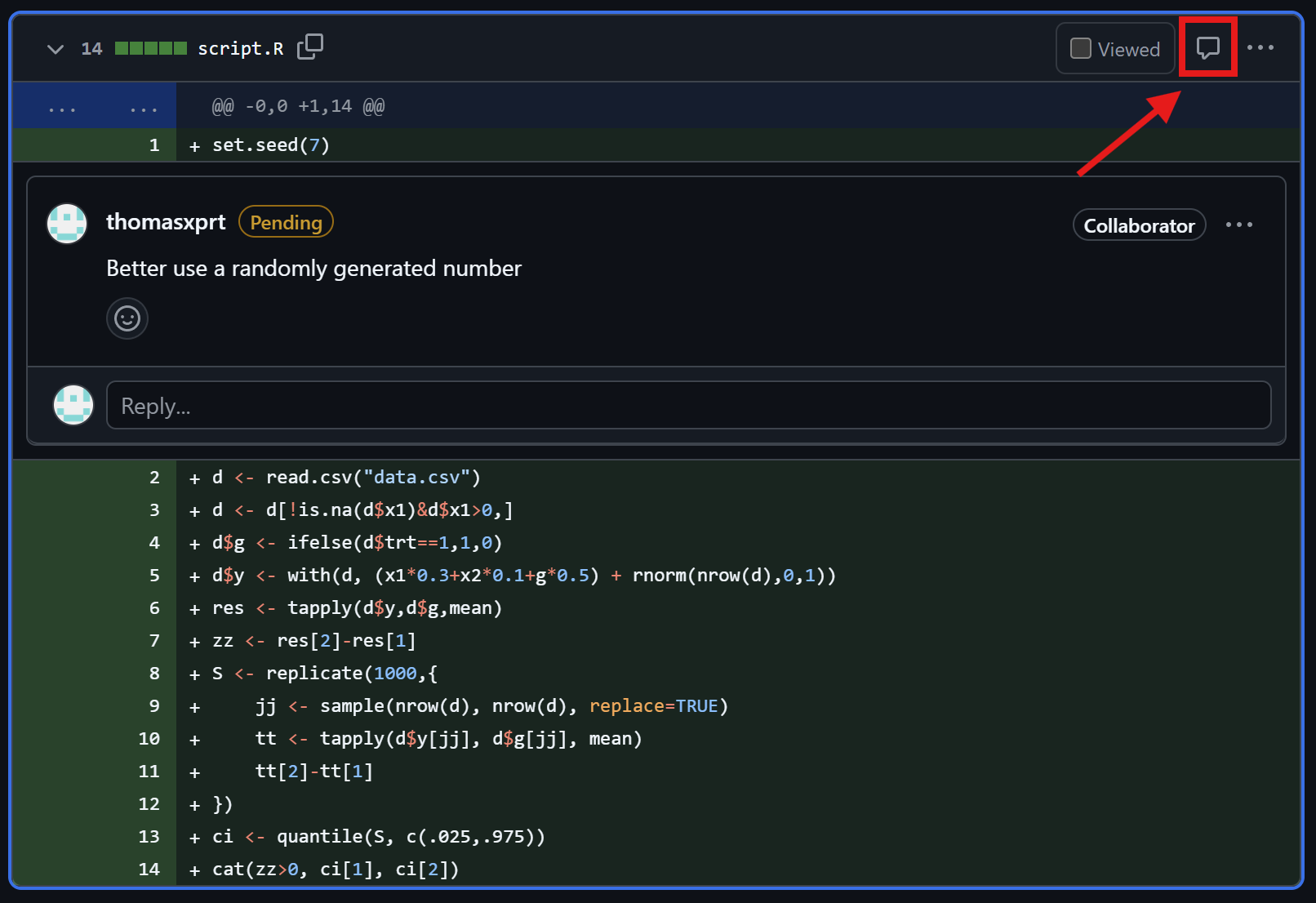
Friedrich checks the comments
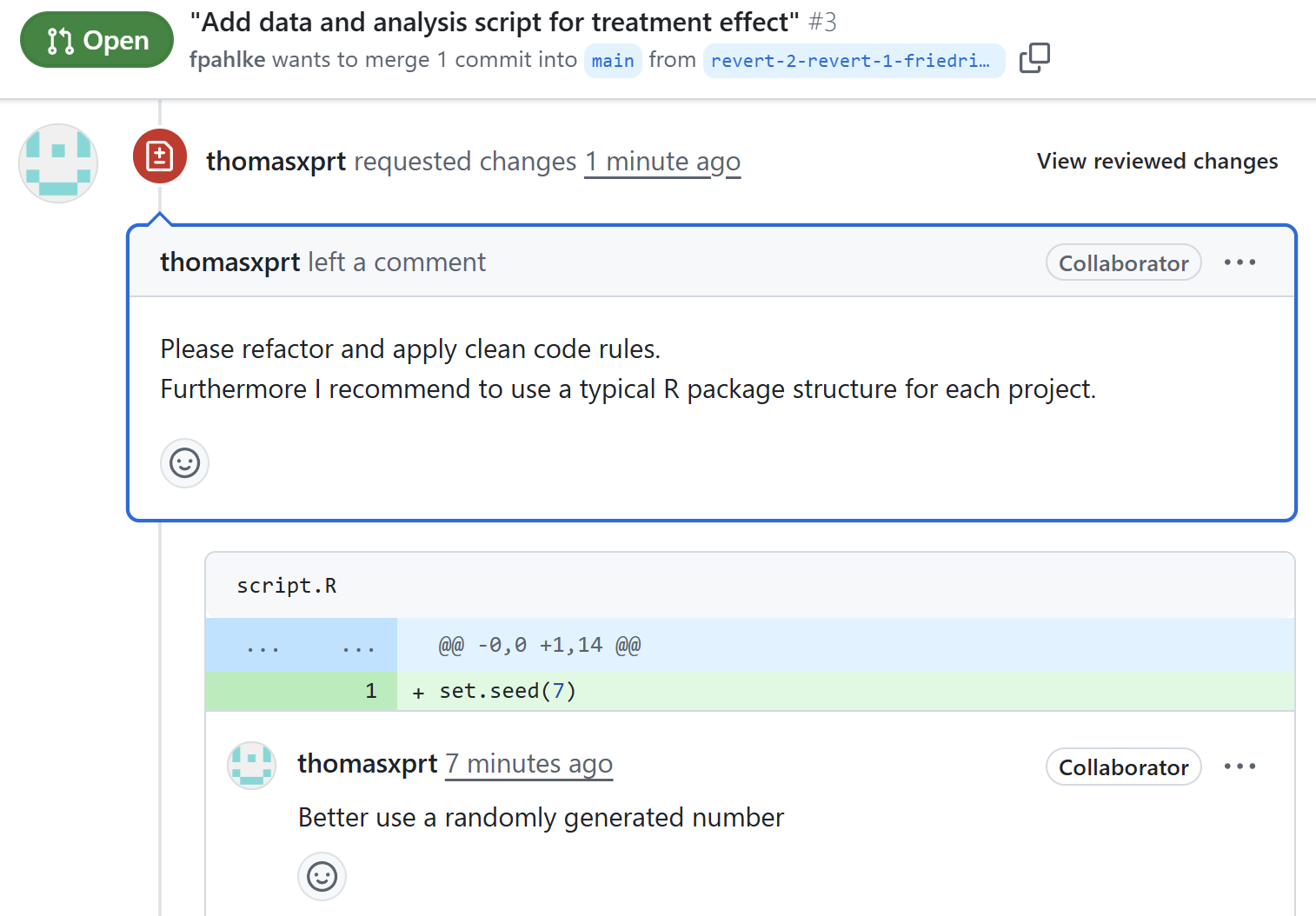
Check reviewer comments
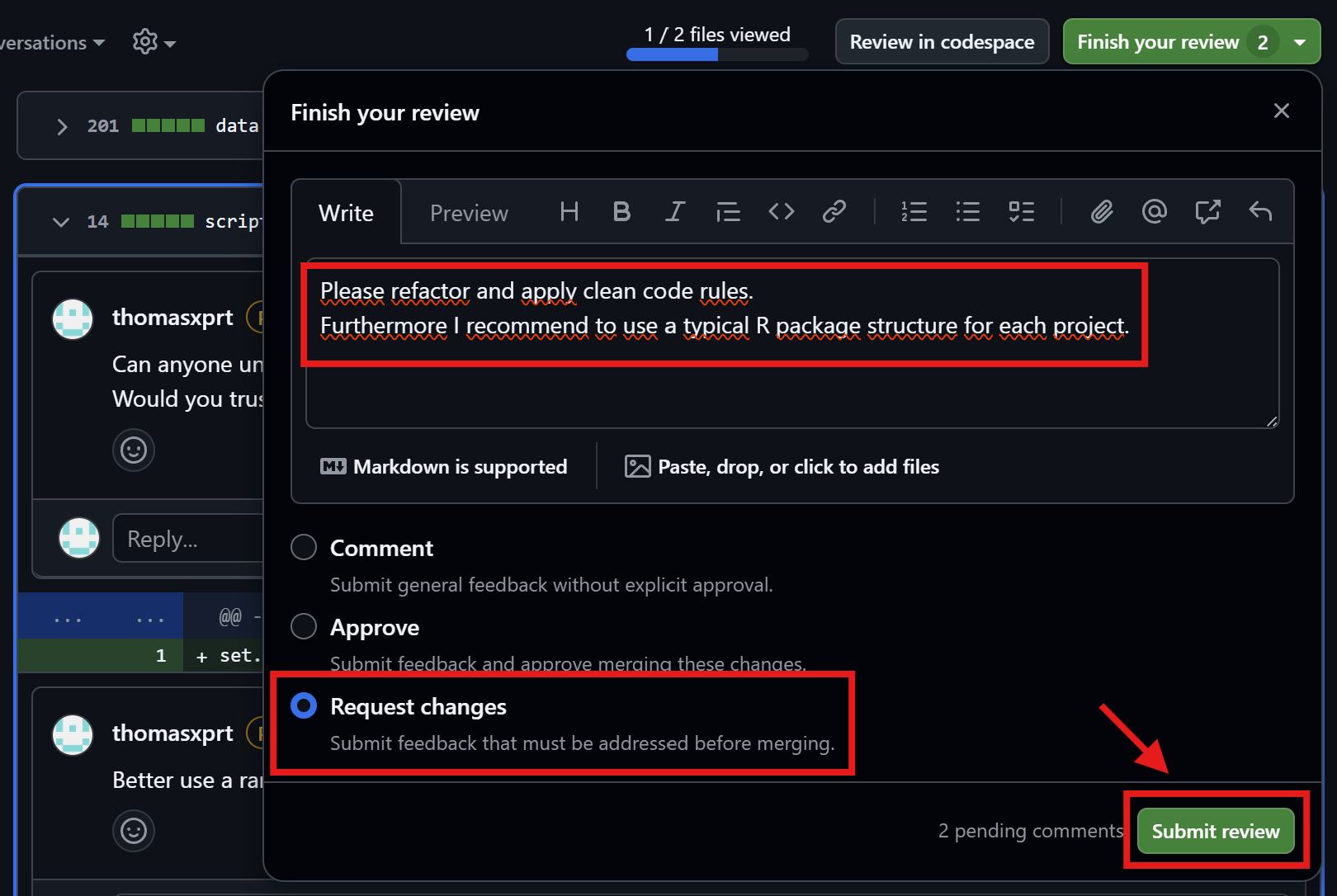
Friedrich fixes the issues
We use the usethis package to create a new R package structure that offers various advantages, even for projects that are not R package projects:
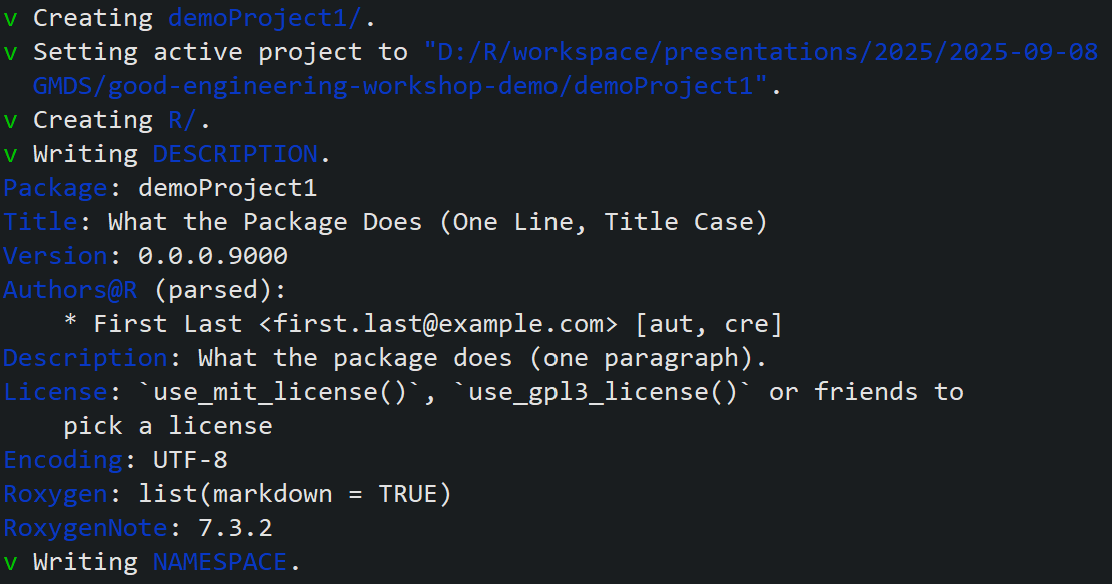
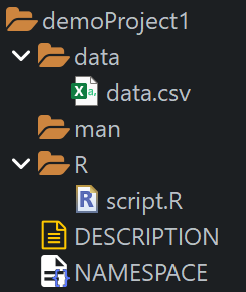
Friedrich commits the changes
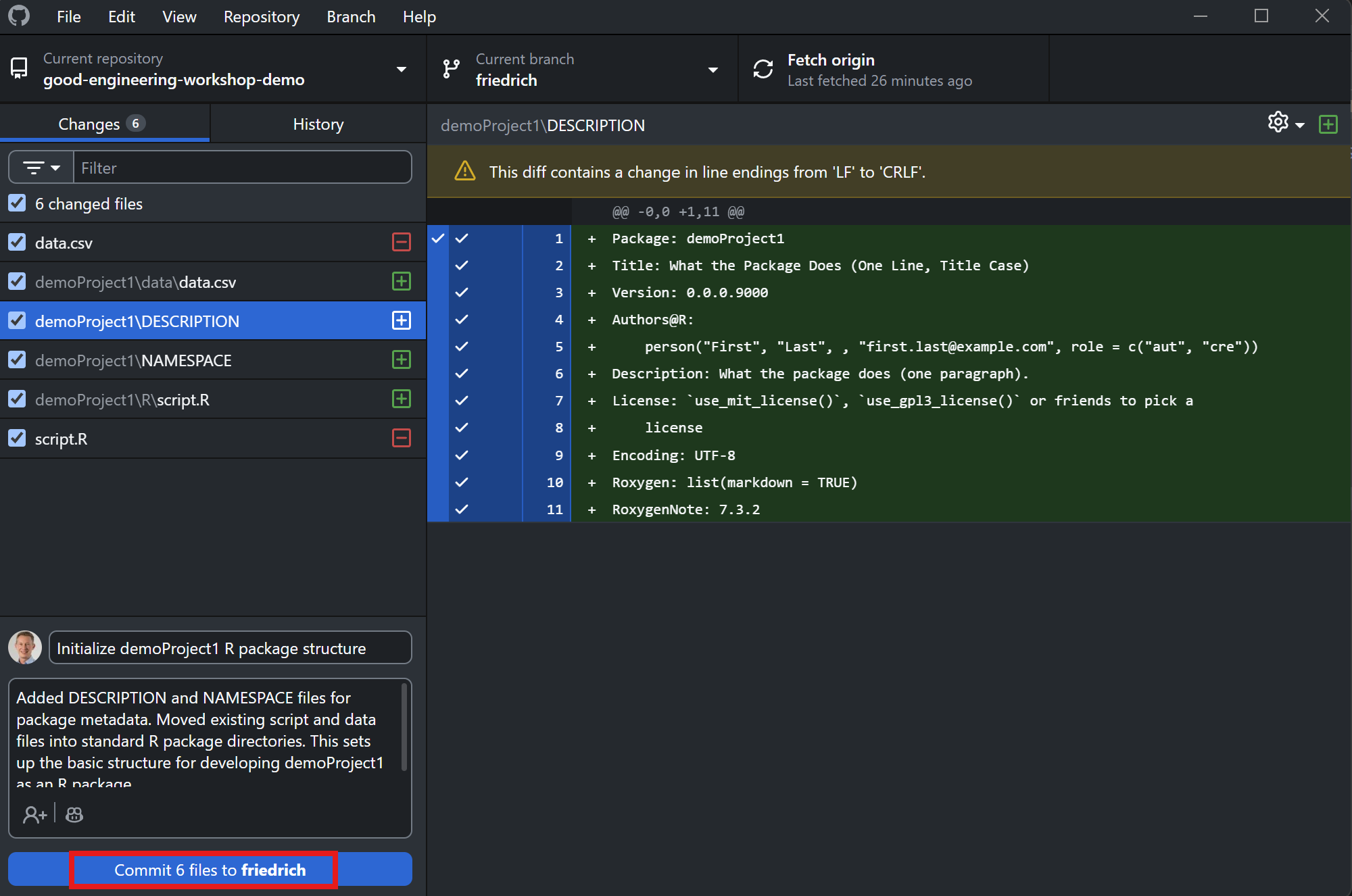
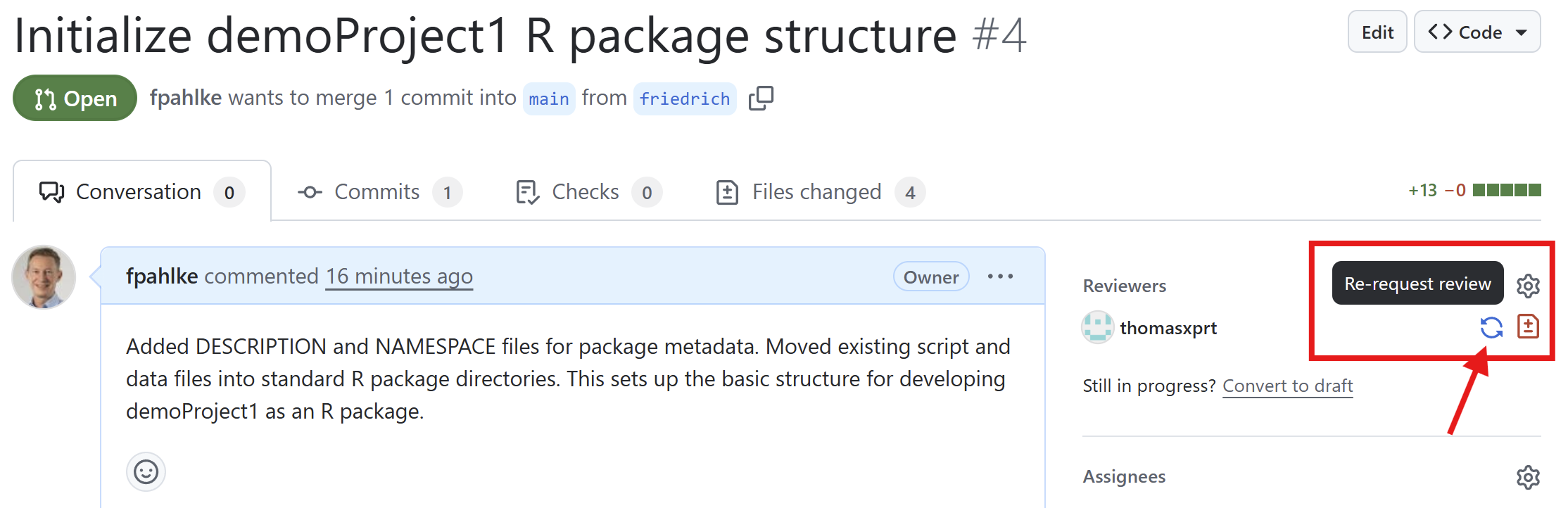
Thomas reviews the changes
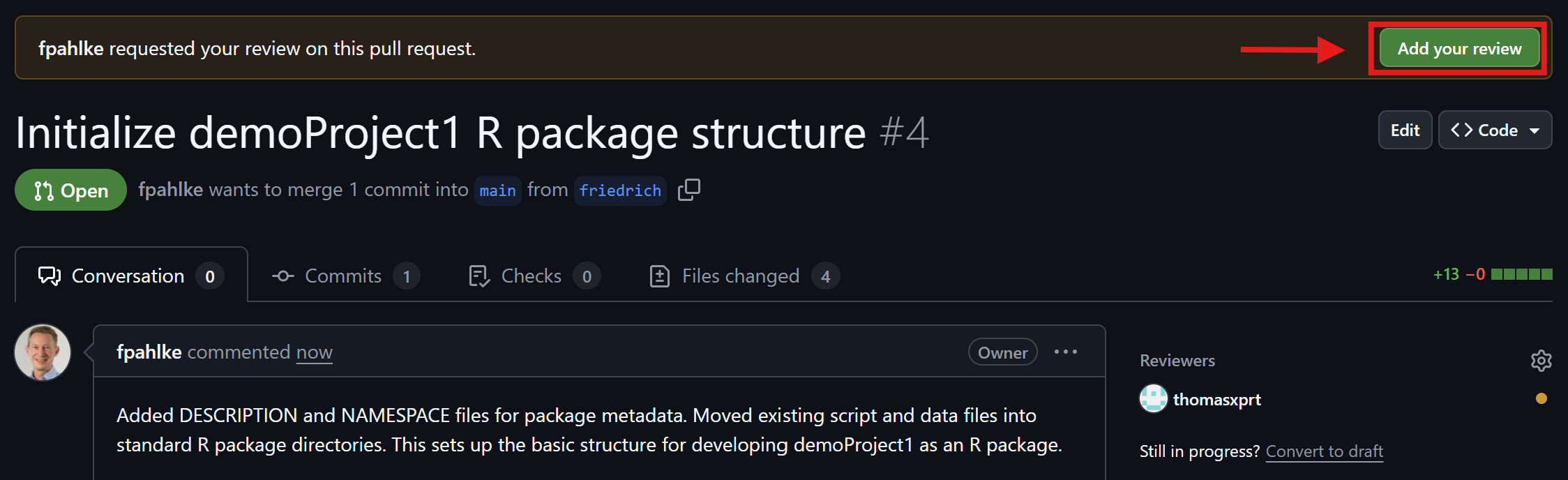

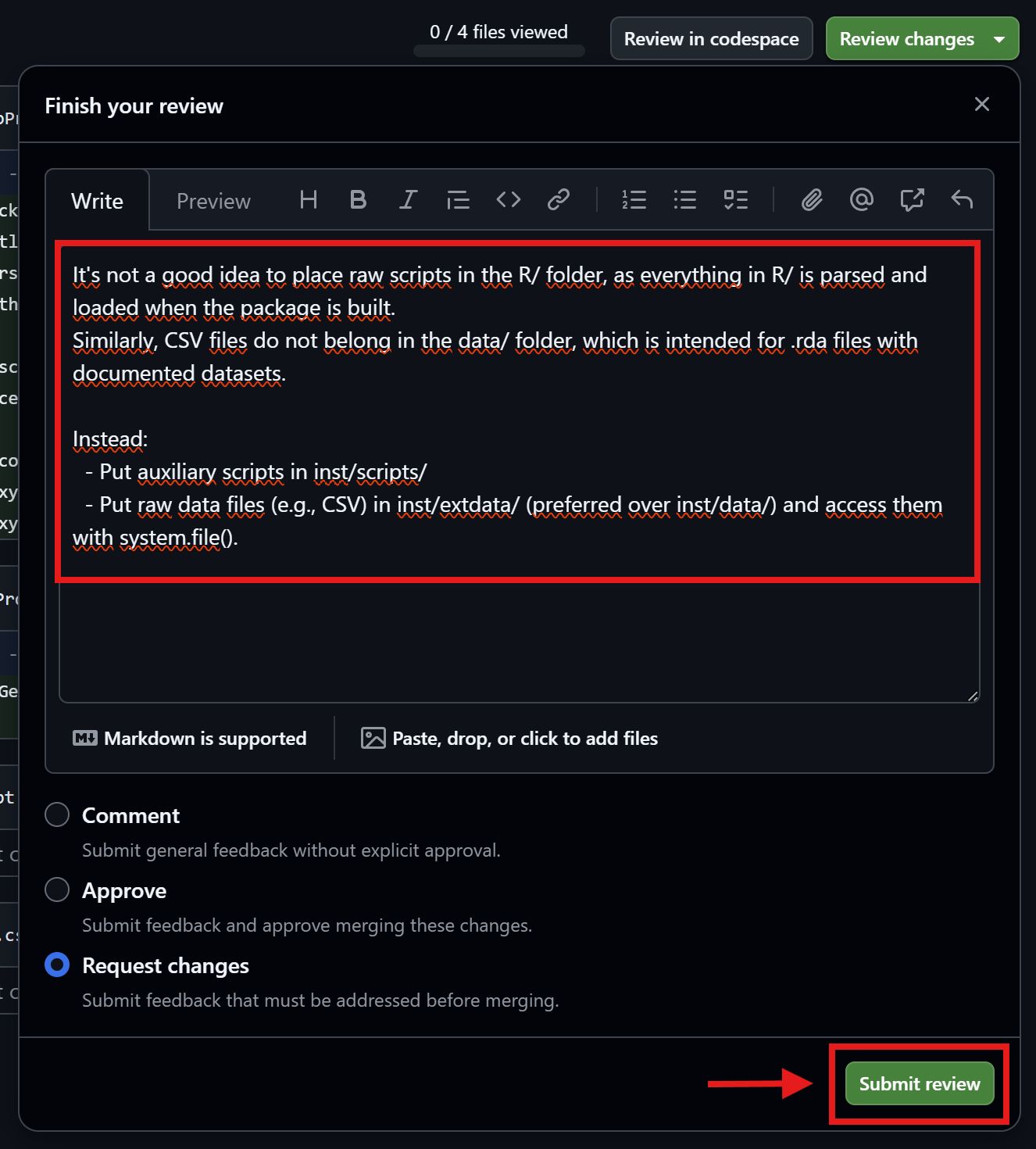
Friedrich checks the comments (2nd round)
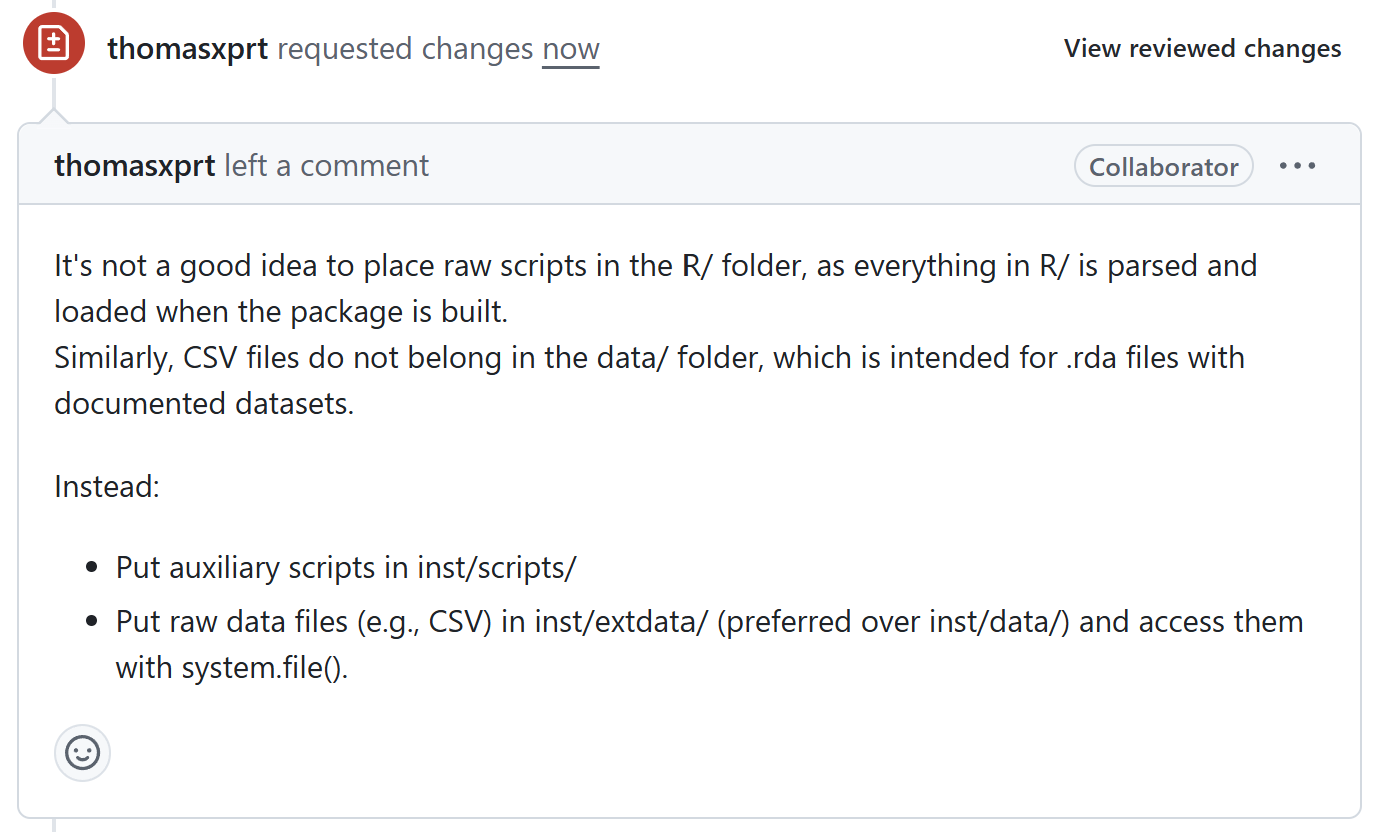
Check reviewer comments
Friedrich fixes the issues (2nd round)
- Put auxiliary scripts in inst/scripts/
- Put raw data files (e.g., CSV) in
inst/extdata/(preferred overinst/data/)
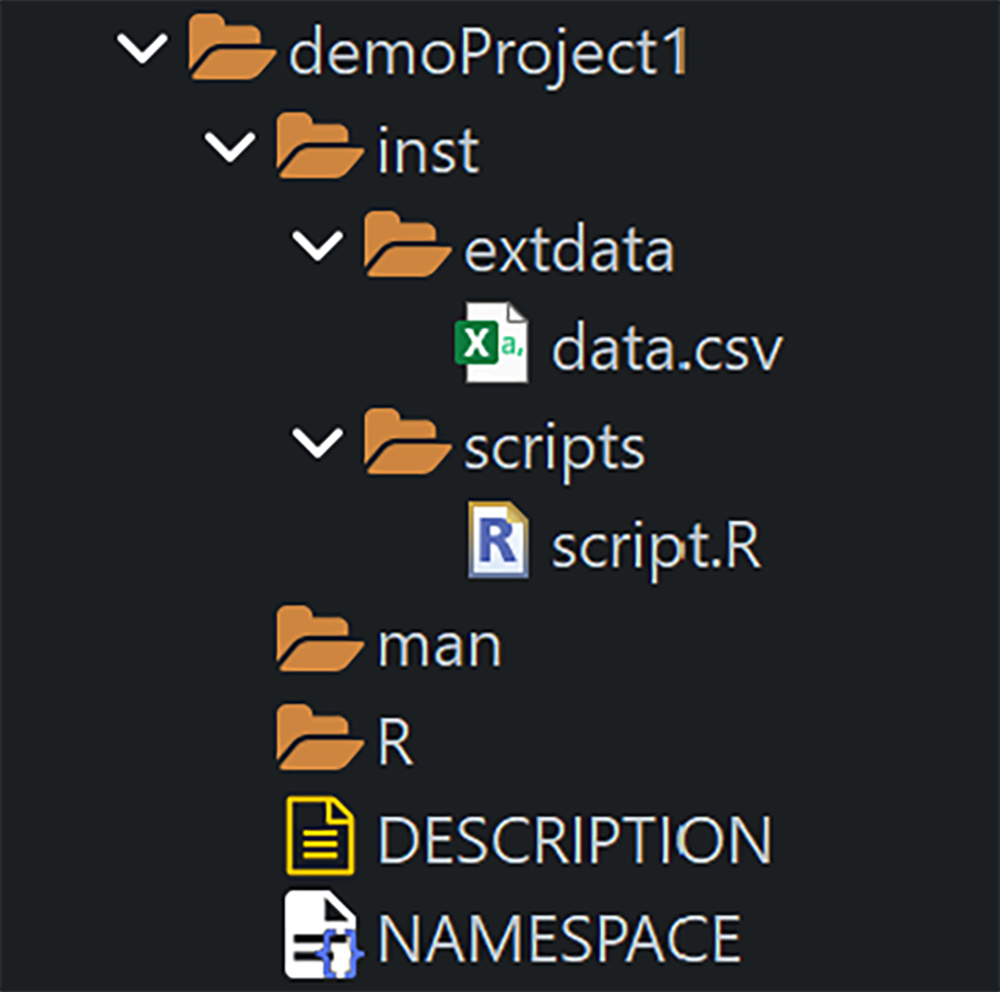
Restructure folders as requested by Thomas
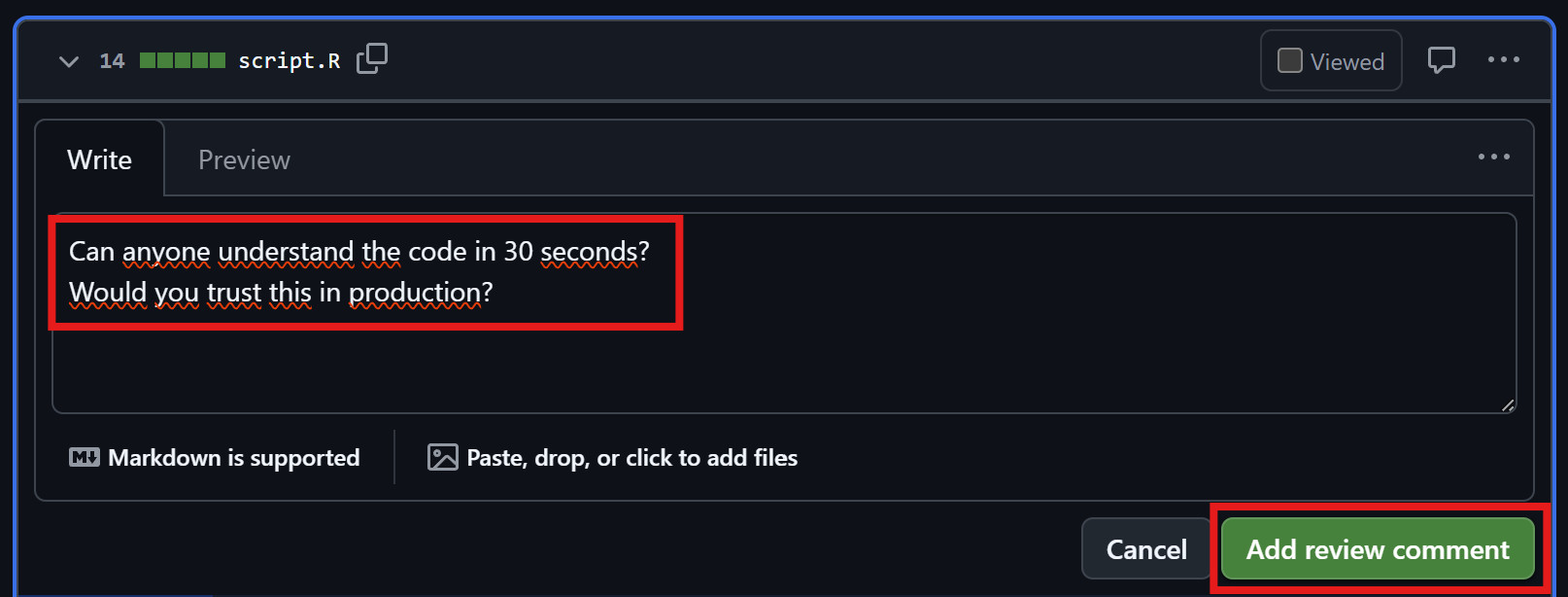
Let’s take a look at script.R: What does this do?
Try to guess in 30 seconds. Would you trust this in production?
set.seed(7)
d <- read.csv("data.csv")
d <- d[!is.na(d$x1)&d$x1>0,]
d$g <- ifelse(d$trt==1,1,0)
d$y <- with(d, (x1*0.3+x2*0.1+g*0.5) + rnorm(nrow(d),0,1))
res <- tapply(d$y,d$g,mean)
zz <- res[2]-res[1]
S <- replicate(1000,{
jj <- sample(nrow(d), nrow(d), replace=TRUE)
tt <- tapply(d$y[jj], d$g[jj], mean)
tt[2]-tt[1]
})
ci <- quantile(S, c(.025,.975))
cat(zz>0, ci[1], ci[2])What’s problematic here?
This script breaks common clean code rules:
- Cryptic names (
d,g,zz,S) - Hidden assumptions (file path, columns exist, coding of
trt) - Mixed responsibilities in one script (load, clean, model, bootstrap, report)
- Magic numbers (
1000,0.025,0.975) - No checks/tests, no explicit output
The same idea — clean Base R version
# Parameters
input_path <- "data.csv"
bootstrap_iterations <- 1000
alpha <- 0.05
seed <- 2486720266 # runif(1, 1e08, 9e08)
# Load & validate
stopifnot(file.exists(input_path))
raw <- read.csv(input_path)
stopifnot(all(c("x1", "x2", "trt") %in% names(raw)))
set.seed(seed)
# Prepare data
prepared <- subset(raw, !is.na(x1) & x1 > 0)
prepared$group <- ifelse(prepared$trt == 1, "treatment", "control")
# Effect estimate function
mean_diff <- function(y, group) {
by_vals <- tapply(y, group, mean)
unname(by_vals["treatment"] - by_vals["control"])
}
# Calculate effect estimate
prepared$y <- with(prepared,
(x1 * 0.3 + x2 * 0.1 + (trt == 1) * 0.5) +
rnorm(nrow(prepared), 0, 1))
estimate <- mean_diff(prepared$y, prepared$group)
# Bootstrap CI
re_idx <- replicate(bootstrap_iterations, sample.int(nrow(prepared),
nrow(prepared), replace = TRUE))
boot_diffs <- apply(re_idx, 2,
function(idx) mean_diff(prepared$y[idx], prepared$group[idx]))
ci <- quantile(boot_diffs,
probs = c(alpha / 2, 1 - alpha / 2),
names = FALSE)
result <- list(estimate = estimate, ci = ci)
resultTidyverse version — even shorter to read
Here, dplyr & friends can improve readability and intent.
# install.packages("dplyr") # if needed
library(dplyr)
params <- list(
input_path = "data.csv",
iterations = 1000,
alpha = 0.05,
seed = 2486720266 # runif(1, 1e08, 9e08)
)
set.seed(params$seed)
raw <- read.csv(params$input_path)
stopifnot(all(c("x1", "x2", "trt") %in% names(raw)))
prepared <- raw |>
filter(!is.na(x1), x1 > 0) |>
mutate(
group = if_else(trt == 1, "treatment", "control"),
y = (x1 * 0.3 + x2 * 0.1 + (trt == 1) * 0.5) + rnorm(n(), 0, 1)
)
mean_diff <- function(df) {
df |>
summarize(diff = mean(y[group == "treatment"]) -
mean(y[group == "control"])) |>
pull(diff)
}
boot_diffs <- replicate(params$iterations, {
s <- sample(nrow(prepared), nrow(prepared), replace = TRUE)
mean_diff(prepared[s, ])
})
ci <- quantile(boot_diffs, c(params$alpha / 2, 1 - params$alpha / 2))
result <- list(estimate = mean_diff(prepared), ci = unname(ci))
resultApply Clean Code Rules

Why is clean code important?
- Maintainability: The code is readable and understandable and has a reduced complexity, i.e., it’s easier to fix bugs
- Extensibility: The architecture is simpler, cleaner, and more expressive, i.e., it’s easier to extend the capabilities and the risk of introducing bugs is reduced
- Performance: The code often runs faster, uses less memory, or is easier to optimize
Why clean code matters (for statisticians)
- Time to result ↓
- Time to handover ↓
- Easier peer review & QA
- Fewer bugs
- Reproducibility & audit readiness (GxP contexts)
- Reusable code ⇒ save time in follow-up projects
- Confidence in outcomes ⇒ better decisions
Example: Clean code rules - Step by step
This script breaks all common clean code rules:
y=function(x){
s1=0
for(v1 in x){s1=s1+v1}
m1=s1/length(x)
i=ceiling(length(x)/2)
if(length(x) %% 2 == 0){i=c(i,i+1)}
s2=0
for(v2 in i){s2=s2+x[v2]}
m2=s2/length(i)
c(m1,m2)
}
y(c(1:7, 100))[1] 16.0 4.5We now refactor it by applying clean code rules…
Example: CCR#1
CCR#1 Naming: Are the names of the variables, functions, and classes descriptive and meaningful?
Naming Conventions: snake_case vs camelCase
- Both are valid — choose based on your context & stay consistent
- snake_case: dominant in R packages developed by Posit, esp. tidyverse style guide
- camelCase: common in several R packages and Base R code, influenced by Java/C#
- Consistency is more important than style choice
Examples:
camelCase eats snake_case
Personal opinion: shorter words, i.e. less to write; as easy to read as snake_case

“Camels may eat snakes to obtain nutrients and cope with their harsh desert environment”
Source: afjrd.org/camels-eating-snakes
Example: CCR#1 — Naming
getMeanAndMedian=function(x){
sum1=0
for(value in x){sum1=sum1+value}
meanValue=sum1/length(x)
centerIndices=ceiling(length(x)/2)
if(length(x) %% 2 == 0){
centerIndices=c(centerIndices,centerIndices+1)
}
sum2=0
for(centerIndex in centerIndices){sum2=sum2+x[centerIndex]}
medianValue=sum2/length(centerIndices)
c(meanValue,medianValue)
}CCR#1 Naming
CCR#2 Formatting: Are indentation, spacing, and bracketing consistent, i.e., is the code easy to read
Example: CCR#2 — Formatting
getMeanAndMedian <- function(x) {
sum1 <- 0
for (value in x) {
sum1 <- sum1 + value
}
meanValue <- sum1 / length(x)
centerIndices <- ceiling(length(x) / 2)
if (length(x) %% 2 == 0) {
centerIndices <- c(
centerIndices, centerIndices + 1)
}
sum2 <- 0
for (centerIndex in centerIndices) {
sum2 <- sum2 + x[centerIndex]
}
medianValue <- sum2 / length(centerIndices)
c(meanValue, medianValue)
}CCR#2 Formatting
CCR#3 Simplicity: Did you keep the code as simple and straightforward as possible, i.e., did you avoid unnecessary complexity
Example: CCR#3 — Simplicity
- From the Simplicity rule also follows: large source files should be split into multiple files
- General guideline: keeping the number of lines to less than 1,000 lines per file can help maintain code readability and manageability
- Put all general and/or reusable functions in the
R/folder - Use descriptive file names., e.g.,
R/load_data.R,R/summarize_parameter.R
- Use
source(list.files(here::here("R"), "\\.R$")to source all R scripts in theR/folder (devtools::load_all()might be useful) - Place calling code in
inst/scripts/(orscripts/), e.g.,inst/scripts/run_analysis.R
Example: CCR#3 — Simplicity
CCR#3 Simplicity
CCR#4 Single Responsibility Principle (SRP): does each function have only a single, well-defined purpose
Example: CCR#4 — Single responsibility principle
getMean <- function(x) {
sum(x) / length(x)
}
isLengthAnEvenNumber <- function(x) {
length(x) %% 2 == 0
}
getMedian <- function(x) {
centerIndices <- ceiling(length(x) / 2)
if (isLengthAnEvenNumber(x)) {
centerIndices <- c(centerIndices, centerIndices + 1)
}
sum(x[centerIndices]) / length(centerIndices)
}CCR#4 Single Responsibility Principle (SRP)
CCR#5 Don’t Repeat Yourself (DRY): Did you avoid duplication of code, either by reusing existing code or creating functions
Example: CCR#5 — DRY
CCR#5: DRY
Suppose you have a code block that performs the same calculation multiple times:
Create a function to encapsulate this calculation and reuse it multiple times:
Example: CCR#5 — DRY
CCR#5 Don’t Repeat Yourself (DRY)
CCR#6 Documentation: Did you use comments to explain the purpose of code blocks and to clarify complex logic
Example: CCR#6 — Documentation
Roxygen (R package roxygen2):
#'
#' Calculate Mean Value
#'
#' @description
#' Computes the arithmetic mean of a numeric vector.
#'
#' @param x A numeric vector.
#'
#' @return A numeric scalar representing the mean of \code{x}.
#'
#' @examples
#' getMean(c(1, 2, 3, 4))
#'
getMean <- function(x) {
sum(x) / length(x)
}
#'
#' Check if Length is Even
#'
#' @description
#' Checks whether the length of the provided vector is even.
#'
#' @param x A vector to check.
#'
#' @return A logical value. Returns \code{TRUE} if the length of
#' \code{x} is even and \code{FALSE} otherwise.
#'
#' @examples
#' isLengthAnEvenNumber(c(1, 2, 3, 4))
#' isLengthAnEvenNumber(1:5)
#'
isLengthAnEvenNumber <- function(x) {
length(x) %% 2 == 0
}
#'
#' Calculate Median
#'
#' @description
#' Computes the median value of a numeric vector.
#' For even-length vectors, the median is calculated
#' as the mean of the two center elements.
#'
#' @param x A numeric vector.
#'
#' @return A numeric scalar representing the median of \code{x}.
#'
#' @examples
#' getMedian(c(1, 3, 5, 7))
#'
getMedian <- function(x) {
centerIndices <- ceiling(length(x) / 2)
if (isLengthAnEvenNumber(x)) {
centerIndices <- c(centerIndices,
centerIndices + 1)
}
getMean(x[centerIndices])
}Example: CCR#6 — Documentation
# returns the mean of x
getMean <- function(x) {
sum(x) / length(x)
}
# returns TRUE if the length of x is
# an even number; FALSE otherwise
isLengthAnEvenNumber <- function(x) {
length(x) %% 2 == 0
}
# returns the median of x
getMedian <- function(x) {
centerIndices <- ceiling(length(x) / 2)
if (isLengthAnEvenNumber(x)) {
centerIndices <- c(centerIndices,
centerIndices + 1)
}
getMean(x[centerIndices])
}Example: CCR#7 — Error handling
#' returns the mean of x
getMean <- function(x) {
checkmate::assertNumeric(x)
sum(x) / length(x)
}
#' returns TRUE if the length of x is an even number; FALSE otherwise
isLengthAnEvenNumber <- function(x) {
checkmate::assertVector(x)
length(x) %% 2 == 0
}
#' returns the median of x
getMedian <- function(x) {
checkmate::assertNumeric(x)
centerIndices <- ceiling(length(x) / 2)
if (isLengthAnEvenNumber(x)) {
centerIndices <- c(centerIndices, centerIndices + 1)
}
getMean(x[centerIndices])
}CCR#7 Error Handling
Summary of Clean Code Rules
- Naming: Use descriptive and meaningful names for variables, functions, and classes
- Formatting: Adhere to consistent indentation, spacing, and bracketing to make the code easy to read
- Simplicity: Keep the code as simple and straightforward as possible, avoiding unnecessary complexity
- Single Responsibility Principle (SRP): Each function should have a single, well-defined purpose
- Don’t Repeat Yourself (DRY): Avoid duplication of code, either by reusing existing code or creating functions
Summary of Clean Code Rules
- Documentation: Use comments to explain the purpose of code blocks and to clarify complex logic
- Error Handling: Include error handling code to gracefully handle exceptions and unexpected situations
- Test-Driven Development (TDD): Write tests for your code to ensure it behaves as expected and to catch bugs early
- Refactoring: Regularly refactor your code to keep it clean, readable, and maintainable
- Code Review: Have other team members review your code to catch potential issues and improve its quality
How to apply Clean Code Rules?
Recommended quality workflow for R scripts and projects:
- Follow the naming and styling guidelines (CCR #1, #2); use tools like styler or Air to automatically format your code
- Continuously write tests and optimize the code coverage with help of tools (CCR #7, #8), especially in GxP contexts
- Document the code and functions (CCR #6); use Roxygen ⇒ HTML documentation can be generated automatically (see pkgdown, GitHub Pages; example: fpahlke.github.io/demoProject1)
- Publish your code on GitHub and invite colleagues to contribute (CCR #10); refactor your code after the review of colleagues and GitHub Copilot (CCR #1, #7, #9)
Testing & Debugging

Use Assertions to check function inputs
- Use assertions inside functions to check input arguments
- Packages like checkmate or assertthat provide many useful assertion functions
Add some sanity tests to your project
R package testthat
- Popular testing framework for R that is easy to learn and use
- Unit testing, integration testing, and snapshot testing supported
- Setup
testthatin your project withusethis::use_testthat()(see below) to create atests/testthat/folder
Example: unit test passed
Example: unit test failed
Error: getMean(c(1, 3, 2, NA)) not equal to 2. Error: getMedian(c(1, 3, 2)) not equal to 2.
Logging & Messages
- Logging is useful for debugging and progress tracking
- Use
message()for progress; keep it short - For larger R scripts, R packages, or Shiny apps:
use a logger package (e.g., loggit, futile.logger, logger, or log4r)
Advantages: log levels, log to file, timestamps, etc.
Reproducibility

Reproducibility essentials
- Use version control (e.g., GitHub)
set.seed()where randomness matters- Record R and R package versions with
sessionInfo() - Use renv for project-level package versions: “A dependency management toolkit for R. Using ‘renv’, you can create and manage project-local R libraries, save the state of these libraries to a ‘lockfile’, and later restore your library as required. Together, these tools can help make your projects more isolated, portable, and reproducible.” (cran.r-project.org/package=renv)
Reproducibility example
Example: sessionInfo()
R version 4.5.1 (2025-06-13)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
time zone: UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] testthat_3.2.3 assertthat_0.2.1
loaded via a namespace (and not attached):
[1] desc_1.4.3 digest_0.6.37 R6_2.6.1 fastmap_1.2.0
[5] xfun_0.53 magrittr_2.0.3 glue_1.8.0 knitr_1.50
[9] htmltools_0.5.8.1 rmarkdown_2.29 lifecycle_1.0.4 cli_3.6.5
[13] vctrs_0.6.5 pkgload_1.4.0 compiler_4.5.1 rprojroot_2.1.1
[17] tools_4.5.1 brio_1.1.5 pillar_1.11.0 evaluate_1.0.5
[21] yaml_2.3.10 rlang_1.1.6 jsonlite_2.0.0 Parameters & Configuration
Avoid the need to edit the source code on different systems and in different repositories, e.g., due to the use of absolute paths.
- Centralize parameters in a JSON or YAML file; usage of .Renviron is also possible
- Use relative paths (e.g., with here)
Parameters in a params.yml file
Use the config package to read the YAML file:
Note: save the yml file in inst/ folder.
R/Quarto Markdown vs Scripts
- R Markdown or Quarto great for exploration, communication, and reporting
- To improve readability, functions should be moved to separate R script files
- Mix them: develop in R Markdown or Quarto, extract clean functions into scripts
- Save R Markdown files in the
vignettes/folder of your project to enable automatic building of documents, reports, or vignettes
(see example project at github.com/fpahlke/demoProject1; easy setup withusethisfunction usethis::use_vignette())
How to optimize the code styling?
Two popular R packages support the tidyverse style guide:
- styler: interactively restyle selected text, files, or entire projects:
- lintr: perform automated checks to confirm that you conform to the style guide
Quite new (2025):
- Air, an extremely fast R formatter
The devtools function spell_check runs a spell check on text fields in the package description file, manual pages, and optionally vignettes.
When tidyverse clearly wins
- Sequence of transformations is linear & readable
- Verbs match your intent (filter, mutate, summarize)
- Fewer temporary objects
library(dplyr)
library(knitr)
data_clean |>
filter(!is.na(y)) |>
mutate(treatment_arm = arm) |>
group_by(treatment_arm) |>
summarize(n = n(),
mean = mean(y),
sd = sd(y),
se = sd(y) / sqrt(length(y))) |>
kable()| treatment_arm | n | mean | sd | se |
|---|---|---|---|---|
| A | 103 | 17.73372 | 8.523611 | 0.8398563 |
| B | 97 | 21.37104 | 6.888219 | 0.6993927 |
Summary

GitHub offers strong benefits
- Even as a small team or solo developer, GitHub offers strong benefits
- Powerful search across all your projects helps you find code quickly
- Keep a clear overview of your work and projects in one place
- Access your repositories securely from anywhere in the world
- Well-maintained
README.mdfiles ensure you still understand your work years later
GitHub for everyday work
- One repo per project; push scripts + outputs (not raw confidential data)
- One branch per developer for clean collaboration
- Meaningful commit messages (Copilot can help)
- Pull requests for code review — combine with GitHub Copilot suggestions
- CI/CD pipelines to automate checks and reporting (GitHub Pages)
Take-home message: GitHub makes everyday work and team collaboration much easier, even in very small teams.
R package structure for projects
Advantages of using an R package structure for projects:
- Built-in documentation
- Easy testing (testthat)
- Dependency management (see DESCRIPTION file and renv)
- GitHub Pages for documentation
- Github Actions for CI/CD
- Easier collaboration: All team members use the same structure and already know where to find things
Example: github.com/fpahlke/demoProject1
LLMs as coding assistants
- Tools like ChatGPT & GitHub Copilot can save hours
- Useful for:
- Generating commit messages for GitHub
- Drafting pull request descriptions
- Reviewing code changes (GitHub PRs ⇒ Copilot)
- Assisting with tricky R/Shiny code
- Writing
roxygen2-style documentation for functions
- Tip: Always review AI-generated code — treat it like a junior colleague’s suggestion
Resources
Example project repository:
- GitHub Repository: github.com/fpahlke/demoProject1
- GitHub Pages: fpahlke.github.io/demoProject1
Example R package repository:
openstatsware working group:
- openstatsguide: Minimum Viable Good Practices for High Quality Statistical Software Packages
Resources (cont’d)
Cloud based coding agents with GitHub integration:
- OpenAI Codex takes on many tasks in parallel, like writing features, answering codebase questions, running tests, and proposing PRs for review. Each task runs in its own secure cloud sandbox, preloaded with your GitHub repository.
- Google Jules tackles bugs, small feature requests, and other software engineering tasks, with direct export to GitHub.
Coding agents for the command line:
- OpenAI Codex CLI is a coding agent that runs locally on your computer in your command line interface.
- Google Gemini CLI: Gemini CLI is an open-source AI agent that provides lightweight access to Gemini, giving you a direct path from your prompt to the Gemini model.
Takeaways
- Code is read more than written — optimise for the reader
- Small, named steps > giant clever one-liners
- Centralize or outsource parameters, validate inputs, set seeds
- Use the typical R package structure for your projects
- Prefer clarity; use tidyverse when it clearly improves readability
- Use the available tools (incl. LLMs) to automate styling, testing, and documentation
Q&A

Your scenarios, your code, …
References
License information
- Creator: Friedrich Pahlke
- This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License
- The slides are hosted at gmds2025.rpact.com
- Important: To use this work you must provide the name of the creators, a link to the material, a link to the license, and indicate if changes were made